 The genetic history of Africa is composed of the overall genetic history of Demographics of Africa, African populations in Africa, including the regional genetic histories of North Africa, West Africa, East Africa, Central Africa, and Southern Africa, as well as the recent African origin of modern humans, recent origin of modern humans in Africa. The Sahara served as a trans-regional passageway and place of dwelling for people in Africa during various Interstadial, humid phases and periods throughout the history of Africa.
The genetic history of Africa is composed of the overall genetic history of Demographics of Africa, African populations in Africa, including the regional genetic histories of North Africa, West Africa, East Africa, Central Africa, and Southern Africa, as well as the recent African origin of modern humans, recent origin of modern humans in Africa. The Sahara served as a trans-regional passageway and place of dwelling for people in Africa during various Interstadial, humid phases and periods throughout the history of Africa.
Overview
 The Demographics of Africa, peoples of Africa are characterized by regional genetic substructure and heterogeneity, depending on the respective ethno-linguistic identity, and, in part, explainable by the "multiregional evolution" of modern human lineages in various multiple regions of the African continent, as well as later admixture events, including back-migrations from Eurasia, of both highly differentiated West- and East-Eurasian components.
Africans' genetic ancestry is largely partitioned by geography and language family, with populations belonging to the same ethno-linguistic groupings showing high genetic homogeneity and coherence. Gene flow, consistent with both short- and long-range migration events followed by extensive admixture and bottleneck effect, bottleneck events, have influenced the regional genetic makeup and demographic structure of Africans. The historical Bantu expansion had lasting impacts on the modern demographic make up of Africa, resulting in a greater genetic and linguistic homogenization. Genetic, archeologic, and linguistic studies added extra insight into this movement: "Our results reveal a genetic continuum of Niger–Congo speaker populations across the continent and extend our current understanding of the routes, timing and extent of the Bantu migration."
Overall, different African populations display genetic diversity and substructure, but can be clustered in distinct but partially overlapping groupings:
* Khoisan hunter-gatherer lineages from Southern Africa represent the deepest lineages, forming a divergent and distinct cluster, shifted away from contemporary "Sub-Saharan Africans", and are further diverged from them than the various Eurasian lineages are. The diverging date of these "Southern hunter-gatherers" from all other human populations is estimated to over 100,000 years ago respectively, with the Khoisan later diverging into two subgroups, northern and southern Khoisan,~30,000 years ago.
* Rain forest foragers such as the Baka people (Cameroon and Gabon), Baka and the Mbuti people, Mbuti diverged from other Sub-Saharan African groups over 60,000 years ago. Eastern groups such as the Mbuti split from Western groups such as the Baka ~20,000 years ago.
* The various Afroasiatic languages, Afroasiatic speakers are suggested to have diverged from other African groups ~50,000 years ago.
* Niger Congo, Niger-Congo and Nilo-Saharan languages, Nilo-Saharan speakers split around 28,000 years ago.
* Austronesian languages, Austronesian-speaking Malagasy people in Madagascar have received significant East Asian people, East/Southeast Asian admixture, less among some groups of coastal Southern, Eastern and the Horn of Africa. The estimated date of geneflow is 2,200 years ago.
The Demographics of Africa, peoples of Africa are characterized by regional genetic substructure and heterogeneity, depending on the respective ethno-linguistic identity, and, in part, explainable by the "multiregional evolution" of modern human lineages in various multiple regions of the African continent, as well as later admixture events, including back-migrations from Eurasia, of both highly differentiated West- and East-Eurasian components.
Africans' genetic ancestry is largely partitioned by geography and language family, with populations belonging to the same ethno-linguistic groupings showing high genetic homogeneity and coherence. Gene flow, consistent with both short- and long-range migration events followed by extensive admixture and bottleneck effect, bottleneck events, have influenced the regional genetic makeup and demographic structure of Africans. The historical Bantu expansion had lasting impacts on the modern demographic make up of Africa, resulting in a greater genetic and linguistic homogenization. Genetic, archeologic, and linguistic studies added extra insight into this movement: "Our results reveal a genetic continuum of Niger–Congo speaker populations across the continent and extend our current understanding of the routes, timing and extent of the Bantu migration."
Overall, different African populations display genetic diversity and substructure, but can be clustered in distinct but partially overlapping groupings:
* Khoisan hunter-gatherer lineages from Southern Africa represent the deepest lineages, forming a divergent and distinct cluster, shifted away from contemporary "Sub-Saharan Africans", and are further diverged from them than the various Eurasian lineages are. The diverging date of these "Southern hunter-gatherers" from all other human populations is estimated to over 100,000 years ago respectively, with the Khoisan later diverging into two subgroups, northern and southern Khoisan,~30,000 years ago.
* Rain forest foragers such as the Baka people (Cameroon and Gabon), Baka and the Mbuti people, Mbuti diverged from other Sub-Saharan African groups over 60,000 years ago. Eastern groups such as the Mbuti split from Western groups such as the Baka ~20,000 years ago.
* The various Afroasiatic languages, Afroasiatic speakers are suggested to have diverged from other African groups ~50,000 years ago.
* Niger Congo, Niger-Congo and Nilo-Saharan languages, Nilo-Saharan speakers split around 28,000 years ago.
* Austronesian languages, Austronesian-speaking Malagasy people in Madagascar have received significant East Asian people, East/Southeast Asian admixture, less among some groups of coastal Southern, Eastern and the Horn of Africa. The estimated date of geneflow is 2,200 years ago.

Indigenous Africans
The Indigenous peoples of Africa, indigenous populations of Africa consists of Niger–Congo languages, Niger–Congo speakers, Nilo-Saharan languages, Nilo-Saharan speakers, the divergent and diverse Khoisan languages, Khoisan grouping, as well as of several unclassified or Language isolate, isolated ethnolinguistic groupings (see :Unclassified languages of Africa, unclassified languages of Africa). The origin of the Afroasiatic languages remains disputed, with some proposing a Middle Eastern origin, while others support an African origin with varying degrees of Eurasian and African components. The Austronesian languages originated in southern East Asia, and later expanded outgoing from the Philippines. The Niger–Congo languages probably originated in or near the area where these languages were spoken prior to Bantu expansion (i.e. West Africa or Central Africa). Its expansion may have been associated with the expansion of agriculture, during in the Neolithic, African Neolithic period, following the African humid period#End, desiccation of the Sahara in c. 3500 BCE. Proto-Niger-Congo may have originated about 10,000 years before present in the "Green Sahara" of Africa (roughly the Sahel and southern Sahara), and that its dispersal can be correlated with the spread of the bow and arrow by migrating hunter-gatherers, which later developed agriculture.
Although the validity of the Nilo-Saharan languages, Nilo-Saharan family remains controversial, the region between Chad, Sudan, and the Central African Republic is seen as a likely candidate for its homeland prior to its dispersal around 10,000–8,000 BCE.
The Southern African hunter-gatherers (Khoisan) are suggested to represent the autochthonous hunter-gatherer population of southern Africa, prior to the expansion of Bantu-speakers from Western/Central Africa and East African pastoralists. Khoisan show evidence for Bantu-related admixture, ranging from nearly ~0% to up to ~87.1%.
The Niger–Congo languages probably originated in or near the area where these languages were spoken prior to Bantu expansion (i.e. West Africa or Central Africa). Its expansion may have been associated with the expansion of agriculture, during in the Neolithic, African Neolithic period, following the African humid period#End, desiccation of the Sahara in c. 3500 BCE. Proto-Niger-Congo may have originated about 10,000 years before present in the "Green Sahara" of Africa (roughly the Sahel and southern Sahara), and that its dispersal can be correlated with the spread of the bow and arrow by migrating hunter-gatherers, which later developed agriculture.
Although the validity of the Nilo-Saharan languages, Nilo-Saharan family remains controversial, the region between Chad, Sudan, and the Central African Republic is seen as a likely candidate for its homeland prior to its dispersal around 10,000–8,000 BCE.
The Southern African hunter-gatherers (Khoisan) are suggested to represent the autochthonous hunter-gatherer population of southern Africa, prior to the expansion of Bantu-speakers from Western/Central Africa and East African pastoralists. Khoisan show evidence for Bantu-related admixture, ranging from nearly ~0% to up to ~87.1%.
Out-of-Africa event

 According to Durvasula et al. (2020), there are indications that 2% to 19% (≃6.6 to 7.0%) of the DNA of West African populations may have come from an unknown archaic hominin which split from the ancestor of humans and Neanderthals between 360 kya to 1.02 mya. However, the study also suggests that at least part of this archaic admixture is also present in Eurasians/non-Africans, and that the admixture event or events range from 0 to 124 ka B.P, which includes the period before the Out-of-Africa migration and prior to the African/Eurasian split (thus affecting in part the common ancestors of both Africans and Eurasians/non-Africans). Chen et al. (2020) found that Africans have higher Neanderthal ancestry than previously thought. 2,504 African samples from all over Africa were analyzed and tested on Neanderthal ancestry. All African samples showed evidence for minor Neanderthal ancestry, but always at lower levels than observed in Eurasians.
According to Durvasula et al. (2020), there are indications that 2% to 19% (≃6.6 to 7.0%) of the DNA of West African populations may have come from an unknown archaic hominin which split from the ancestor of humans and Neanderthals between 360 kya to 1.02 mya. However, the study also suggests that at least part of this archaic admixture is also present in Eurasians/non-Africans, and that the admixture event or events range from 0 to 124 ka B.P, which includes the period before the Out-of-Africa migration and prior to the African/Eurasian split (thus affecting in part the common ancestors of both Africans and Eurasians/non-Africans). Chen et al. (2020) found that Africans have higher Neanderthal ancestry than previously thought. 2,504 African samples from all over Africa were analyzed and tested on Neanderthal ancestry. All African samples showed evidence for minor Neanderthal ancestry, but always at lower levels than observed in Eurasians.
Geneflow between Eurasian and African populations
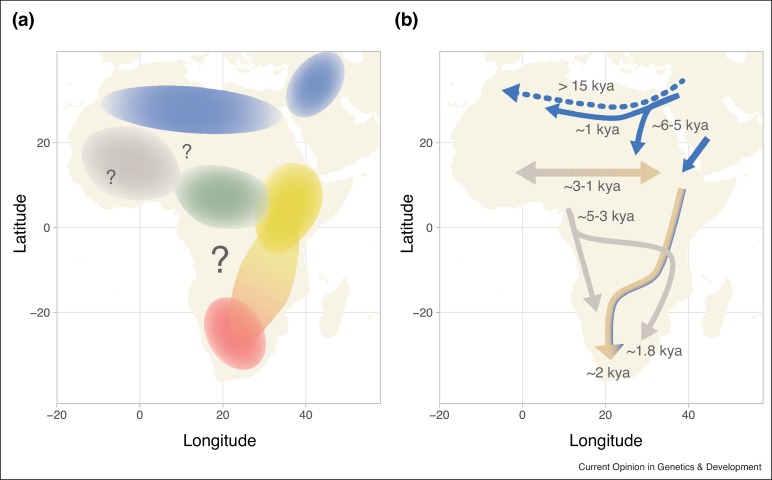 Significant Eurasian admixture is found in Northern Africa, and among specific ethnic groups of the Horn of Africa, as well as among the Malagasy people of Madagascar. Various genome studies found evidence for multiple Prehistory, prehistoric back-migrations from various Eurasian populations and subsequent admixture with native groups. West-Eurasian geneflow arrived to Northern Africa during the Paleolithic (30,000 to 15,000 years ago), followed by other pre-Neolithic and Neolithic migration events. Genetic data on the Taforalt samples "demonstrated that Northern Africa received significant amounts of gene-flow from Eurasia predating the Holocene and development of farming practices". Medieval geneflow events, such as the Arab expansion also left traces in various African populations. Pickrell et al. (2014) indicated that Western Eurasian ancestry eventually arrived through Northeast Africa (particularly the Horn of Africa) to Southern Africa.
Ramsay et al. (2018) also found evidence for significant Western Eurasian admixture in various parts of Africa, from both ancient and more recent migrations, being highest among populations from North Africa, Northern Africa, and some groups of the Horn of Africa:
Significant Eurasian admixture is found in Northern Africa, and among specific ethnic groups of the Horn of Africa, as well as among the Malagasy people of Madagascar. Various genome studies found evidence for multiple Prehistory, prehistoric back-migrations from various Eurasian populations and subsequent admixture with native groups. West-Eurasian geneflow arrived to Northern Africa during the Paleolithic (30,000 to 15,000 years ago), followed by other pre-Neolithic and Neolithic migration events. Genetic data on the Taforalt samples "demonstrated that Northern Africa received significant amounts of gene-flow from Eurasia predating the Holocene and development of farming practices". Medieval geneflow events, such as the Arab expansion also left traces in various African populations. Pickrell et al. (2014) indicated that Western Eurasian ancestry eventually arrived through Northeast Africa (particularly the Horn of Africa) to Southern Africa.
Ramsay et al. (2018) also found evidence for significant Western Eurasian admixture in various parts of Africa, from both ancient and more recent migrations, being highest among populations from North Africa, Northern Africa, and some groups of the Horn of Africa:
In addition to the intrinsic diversity within the continent due to population structure and isolation, migration of Eurasian populations into Africa has emerged as a critical contributor to the genetic diversity. These migrations involved the influx of different Eurasian populations at different times and to different parts of Africa. Comprehensive characterization of the details of these migrations through genetic studies on existing populations could help to explain the strong genetic differences between some geographically neighbouring populations. This distinctive Eurasian admixture appears to have occurred over at least three time periods with ancient admixture in central west Africa (e.g., Yoruba from Nigeria) occurring between ~7.5 and 10.5 kya, older admixture in east Africa (e.g., Ethiopia) occurring between ~2.4 and 3.2 kya and more recent admixture between ~0.15 and 1.5 kya in some east African (e.g., Kenyan) populations. Subsequent studies based on LD decay and haplotype sharing in an extensive set of African and Eurasian populations confirmed the presence of Eurasian signatures in west, east and southern Africans. In the west, in addition to Niger-Congo speakers from The Gambia and Mali, the Mossi from Burkina Faso showed the oldest Eurasian admixture event ~7 kya. In the east, these analyses inferred Eurasian admixture within the last 4000 years in Kenya.
 While many studies conducted on Horn of Africa populations estimate a West-Eurasian admixture event around 3,000 years ago, Hodgson et al. (2014) found a distinct West-Eurasian ancestral component among studied Afroasiatic-speaking groups in the Horn of Africa (and to a lesser extent in North Africa and Western Asia, West Asia), most prevalent among the Somalis, Somali. This ancestral component — dubbed "Ethio-Somali" — would have diverged from other Non-Africans (genetic lineage), non-African ancestries around 23,000 years ago and migrated back to Africa prior to developing agriculture, merging with the local indigenous lineages of the Horn of Africa. The authors propose that "Ethio-Somali" may have been a substantial ancestral component of the Proto-Afroasiatic-speaking population. A subsequent Mitochondrial DNA, mtDNA analysis by Gandini et al. (2016) has produced additional evidence in support of a pre-agricultural back-migration from West-Eurasia into the Horn of Africa with an estimated date of arrival into the Horn of Africa in the early Holocene, possibly as a result of obsidian exchange networks across the Red Sea. Hodgson et al. also confirmed the existence of an ancestral component indigenous to the Horn of Africa – "Ethiopic" (Hodgson et al.) or "Omotic" (Pagani et al.) – which is most prevalent among speakers of the Omotic languages, Omotic branch of Afroasiatic in southwestern Ethiopia. This lineage is associated with that of a 4,500 year-old fossil (Mota) found in a cave in southwestern Ethiopia, which has high genetic affinity to modern Ethiopians, Ethiopian groups, especially the Endogamy, endogamous blacksmith caste of the Omotic Ari people, Aari people. Like Mota, Aari blacksmiths do not show evidence for admixture with West-Eurasians, demonstrating a degree of population continuity in this region for at least 4,500 years. In a comparative analysis of Mota’s genome referencing modern populations, Gallego et al. (2016) concluded that the divergence of Omotic from other Afroasiatic languages may have resulted from the relative isolation of its speakers from external groups. In an analysis of 68 Ethiopian ethnic groups, also referencing Mota, Lopez et al. (2021) revealed that groups belonging to the Omotic, Cushitic, and Semitic branches of Afro-Asiatic show high genetic similarity to each other on average. The data also support widespread recent intermixing among ethnic groups. Linguist Roger Blench proposed southwestern Ethiopia as the most likely homeland of Afroasiatic, due in part to the high internal diversification of the Omotic branch spoken in that region. In addition, Human Y-chromosome DNA haplogroup, Y-haplogroup sub-lineage Haplogroup E-M215 (Y-DNA), E-M215 (also known as "E1b1b) and its derivative E-M35 are quite common among Afroasiatic speakers and southwestern Ethiopia is a plausible source of these haplogroups. The linguistic group and carriers of this lineage have a high probability to have arisen and dispersed together from Northeast Africa in the Mesolithic, plausibly having already developed subsistence patterns of pastoralism and intensive plant usage and collection. According to historian and linguist Christopher Ehret, the form of intensive plant collection practiced by the Proto-Afroasiatic population in Northeast Africa may have been a precursor to the agricultural practices that would later independently develop in the Fertile Crescent and the Horn of Africa. A 2018 re-analysis of autosomal DNA using modern populations as a reference found that the ancient Natufian samples of the Levant harbored 6.8% Omotic-related ancestry. Dobon et al. (2015) identified an autosomal ancestral component that is commonly found among modern Afroasiatic-speaking populations (as well as Nubians) in Northeast Africa. This component, which peaks among Copts in Sudan but is not found in Egyptians or Qataris, appears alongside a component that defines Nilo-Saharan languages, Nilo-Saharan speakers of southwestern Sudan and South Sudan. Arauna et al. (2017), analyzing existing genetic data obtained from North Africa, Northern African populations, such as Berbers, described them as a mosaic of Middle Eastern, European, and Sub-Saharan African-related ancestries.
Specific East Asian-related ancestry is found among the Malagasy languages, Malagasy speakers of Madagascar at a medium frequency. The presence of this East Asian-related ancestry is mostly linked to the Austronesian peoples expansion from Southeast Asia. The peoples of Borneo were identified to resemble the East Asian voyagers, who arrived on Madagascar. East Asian ancestry among Malagasy people was estimated at a mean average of 33%, but as high as ~75% among some Highlander groups and upper caste groups.
Chen et al. (2020) analyzed 2,504 African samples from all over Africa, and found archaic Neanderthal ancestry, among all tested African samples at low frequency. They also identified a European-related (West-Eurasian) ancestry segment, which seems to largely correspond with the detected Neanderthal ancestry components. European-related admixture among Africans was estimated to be between ~0% to up to ~30%, with a peak among Northern Africans. According to Chen et al. (2020), "These data are consistent with the hypothesis that back-migration contributed to the signal of Neanderthal ancestry in Africans. Furthermore, the data indicates that this back-migration came after the split of Europeans and East Asians, from a population related to the European lineage."
Hollfelder et al. (2021) concluded that West African Yoruba people, which were previously used as "unadmixed reference population" for indigenous Africans, harbor minor levels of Neanderthal ancestry, which can be largely associated with back-migration of an "Ancestral European-like" source population.
Multiple studies found also evidence for geneflow of indigenous African ancestry towards Eurasia, specifically Europe and the Middle East. The analysis of 40 different West-Eurasian populations found African admixture at a frequency of 0% to up to ~15%.
There is a minor geneflow from North Africa in parts of Southern Europe, this is supported by the presence of an African-specific mitochondrial haplogroup among one of four 4,000 year old samples.
While many studies conducted on Horn of Africa populations estimate a West-Eurasian admixture event around 3,000 years ago, Hodgson et al. (2014) found a distinct West-Eurasian ancestral component among studied Afroasiatic-speaking groups in the Horn of Africa (and to a lesser extent in North Africa and Western Asia, West Asia), most prevalent among the Somalis, Somali. This ancestral component — dubbed "Ethio-Somali" — would have diverged from other Non-Africans (genetic lineage), non-African ancestries around 23,000 years ago and migrated back to Africa prior to developing agriculture, merging with the local indigenous lineages of the Horn of Africa. The authors propose that "Ethio-Somali" may have been a substantial ancestral component of the Proto-Afroasiatic-speaking population. A subsequent Mitochondrial DNA, mtDNA analysis by Gandini et al. (2016) has produced additional evidence in support of a pre-agricultural back-migration from West-Eurasia into the Horn of Africa with an estimated date of arrival into the Horn of Africa in the early Holocene, possibly as a result of obsidian exchange networks across the Red Sea. Hodgson et al. also confirmed the existence of an ancestral component indigenous to the Horn of Africa – "Ethiopic" (Hodgson et al.) or "Omotic" (Pagani et al.) – which is most prevalent among speakers of the Omotic languages, Omotic branch of Afroasiatic in southwestern Ethiopia. This lineage is associated with that of a 4,500 year-old fossil (Mota) found in a cave in southwestern Ethiopia, which has high genetic affinity to modern Ethiopians, Ethiopian groups, especially the Endogamy, endogamous blacksmith caste of the Omotic Ari people, Aari people. Like Mota, Aari blacksmiths do not show evidence for admixture with West-Eurasians, demonstrating a degree of population continuity in this region for at least 4,500 years. In a comparative analysis of Mota’s genome referencing modern populations, Gallego et al. (2016) concluded that the divergence of Omotic from other Afroasiatic languages may have resulted from the relative isolation of its speakers from external groups. In an analysis of 68 Ethiopian ethnic groups, also referencing Mota, Lopez et al. (2021) revealed that groups belonging to the Omotic, Cushitic, and Semitic branches of Afro-Asiatic show high genetic similarity to each other on average. The data also support widespread recent intermixing among ethnic groups. Linguist Roger Blench proposed southwestern Ethiopia as the most likely homeland of Afroasiatic, due in part to the high internal diversification of the Omotic branch spoken in that region. In addition, Human Y-chromosome DNA haplogroup, Y-haplogroup sub-lineage Haplogroup E-M215 (Y-DNA), E-M215 (also known as "E1b1b) and its derivative E-M35 are quite common among Afroasiatic speakers and southwestern Ethiopia is a plausible source of these haplogroups. The linguistic group and carriers of this lineage have a high probability to have arisen and dispersed together from Northeast Africa in the Mesolithic, plausibly having already developed subsistence patterns of pastoralism and intensive plant usage and collection. According to historian and linguist Christopher Ehret, the form of intensive plant collection practiced by the Proto-Afroasiatic population in Northeast Africa may have been a precursor to the agricultural practices that would later independently develop in the Fertile Crescent and the Horn of Africa. A 2018 re-analysis of autosomal DNA using modern populations as a reference found that the ancient Natufian samples of the Levant harbored 6.8% Omotic-related ancestry. Dobon et al. (2015) identified an autosomal ancestral component that is commonly found among modern Afroasiatic-speaking populations (as well as Nubians) in Northeast Africa. This component, which peaks among Copts in Sudan but is not found in Egyptians or Qataris, appears alongside a component that defines Nilo-Saharan languages, Nilo-Saharan speakers of southwestern Sudan and South Sudan. Arauna et al. (2017), analyzing existing genetic data obtained from North Africa, Northern African populations, such as Berbers, described them as a mosaic of Middle Eastern, European, and Sub-Saharan African-related ancestries.
Specific East Asian-related ancestry is found among the Malagasy languages, Malagasy speakers of Madagascar at a medium frequency. The presence of this East Asian-related ancestry is mostly linked to the Austronesian peoples expansion from Southeast Asia. The peoples of Borneo were identified to resemble the East Asian voyagers, who arrived on Madagascar. East Asian ancestry among Malagasy people was estimated at a mean average of 33%, but as high as ~75% among some Highlander groups and upper caste groups.
Chen et al. (2020) analyzed 2,504 African samples from all over Africa, and found archaic Neanderthal ancestry, among all tested African samples at low frequency. They also identified a European-related (West-Eurasian) ancestry segment, which seems to largely correspond with the detected Neanderthal ancestry components. European-related admixture among Africans was estimated to be between ~0% to up to ~30%, with a peak among Northern Africans. According to Chen et al. (2020), "These data are consistent with the hypothesis that back-migration contributed to the signal of Neanderthal ancestry in Africans. Furthermore, the data indicates that this back-migration came after the split of Europeans and East Asians, from a population related to the European lineage."
Hollfelder et al. (2021) concluded that West African Yoruba people, which were previously used as "unadmixed reference population" for indigenous Africans, harbor minor levels of Neanderthal ancestry, which can be largely associated with back-migration of an "Ancestral European-like" source population.
Multiple studies found also evidence for geneflow of indigenous African ancestry towards Eurasia, specifically Europe and the Middle East. The analysis of 40 different West-Eurasian populations found African admixture at a frequency of 0% to up to ~15%.
There is a minor geneflow from North Africa in parts of Southern Europe, this is supported by the presence of an African-specific mitochondrial haplogroup among one of four 4,000 year old samples.
Regional genomic overview
North Africa
Archaic Human DNA
While Denisovan and Neanderthal ancestry in Non-Africans (genetic lineage), non-Africans outside of Africa are more certain, archaic human ancestry in Africans is less certain and is too early to be established with certainty.Ancient DNA
An early medieval sample from the 5th to 6th century CE found in northern France (Angers) was found to be similar to present-day North Africans.=Egypt
= Tomb of Two Brothers#The tomb owners, Khnum-aa, Tomb of Two Brothers, Khnum-Nakht, Tomb of Two Brothers#Coffins of Nakht-Ankh, Nakht-Ankh and JK2911 carried maternal Haplogroup M (mtDNA)#Haplogroup M1, haplogroup M1a1. Djehutynakht (10A) carried maternal haplogroup Haplogroup U (mtDNA)#Haplogroup U5, U5b2b5. JK2888 carried maternal haplogroup Haplogroup U (mtDNA), U6a2. Thuya, Tiye, Tutankhamen's mother, and Tutankhamen carried the maternal Haplogroup K (mtDNA), haplogroup K. JK2134 carried maternal haplogroup Haplogroup J (mtDNA), J1d and JK2887 carried maternal haplogroup Haplogroup J (mtDNA), J2a1a1. Amenhotep III, Akhenaten, and Tutankhamen carried the paternal haplogroup R1b. Ramesses III and "Unknown Man E", possibly Pentawere, carried paternal haplogroup E1b1a. JK2134 and JK2911 carried paternal haplogroup Haplogroup J (Y-DNA), J. Takabuti carried maternal haplogroup Haplogroup H (mtDNA), H4a1 and YM:KMM A 63 carried maternal haplogroup Haplogroup HV (mtDNA), HV. OM:KMM A 64 carried maternal haplogroup Haplogroup T (mtDNA), T2c1a. JK2888 carried paternal haplogroup Haplogroup E-M215 (Y-DNA), E1b1b1a1b2.=Libya
= At Takarkori rockshelter, in Libya, two naturally mummified women, dated to the Pastoral Period#Middle Pastoral Period 2, Middle Pastoral Period (7000 BP), carried Basal (phylogenetics), basal maternal Haplogroup N (mtDNA), haplogroup N.=Morocco
= Van de Loorsdrecht et al. (2018) found that of seven samples of Taforalts of Morocco, radiocarbon dated to between 15,100 cal BP and 13,900 cal BP, six were found to carry maternal haplogroup Haplogroup U (mtDNA), U6a, and one was found to carry maternal haplogroup Haplogroup M (mtDNA), M1b. Six of six males were found to carry paternal haplogroup Haplogroup E-M215 (Y-DNA), E1b1b. They were found to harbor 63.5% Natufian-related ancestry and 36.5% Sub-Saharan African-related ancestry. The Sub-Saharan component is most strongly drawn out by modern West African groups such as the Yoruba people, Yoruba and the Mende people, Mende. The samples also contain an additional affinity to South, Central, and East African outgroups that cannot be explained by any known ancient or modern populations. When projected onto a principal component analysis graph of African and west Eurasian populations, the Taforalt individuals form a distinct cluster in an intermediate position between present-day North Africans [e.g., Berbers, Amazighes (Berbers), Mozabites, and Saharawis] and East Africans (e.g., Afar people, Afars, Oromos, and Somalis). Jeong (2020), comparing the Taforalt people of the Iberomaurusian culture to modern populations, found that the Taforalt's Sub-Saharan African genetic component may be best represented by modern West Africans (e.g., Yoruba).Y-Chromosomal DNA
Mitochondrial DNA
Amid the Holocene, including the Holocene Climate Optimum in 8000 BP, Africans bearing haplogroup L2 spread within West Africa and Africans bearing haplogroup L3 spread within East Africa. As the largest migration since the Out of Africa theory, Out of Africa migration, migration from Sub-Saharan Africa toward the North Africa occurred, by West Africans, Central Africans, and East Africans, resulting in migrations into Europe and Asia; consequently, Sub-Saharan African mitochondrial DNA was introduced into Europe and Asia. During the early period of the Holocene, 50% of Macro-haplogroup L (mtDNA), Sub-Saharan African mitochondrial DNA was introduced into North Africa by West Africa#Demographics and languages, West Africans and the other 50% was introduced by East Africa#Demographics, East Africans. During the modern period, a greater number of West Africans introduced Sub-Saharan African mitochondrial DNA into North Africa than East Africans. Mitochondrial haplogroups L3, M, and N are found among Sudan#Ethnic groups, Sudanese peoples (e.g., Beja people, Beja, Nilotic peoples, Nilotics, Nuba, Nubians), who have no known interaction (e.g., history of migration/admixture) with Europeans or Asians; rather than having developed in a post-Out-of-Africa migration context, mitochondrial macrohaplogroup L3/M/N and its subsequent development into distinct mitochondrial haplogroups (e.g., Haplogroup L3, Haplogroup M (mtDNA), Haplogroup M, Haplogroup N (mtDNA), Haplogroup N) may have occurred in East Africa at a time that considerably predates the Out-of-Africa migration event of 50,000 BP.Autosomal DNA
Medical DNA
=Lactase Persistence
= Neolithic agriculturalists, who may have resided in Northeast Africa and the Near East, may have been the source population for lactase persistence variants, including –13910*T, and may have been subsequently supplanted by later migrations of peoples. The Sub-Saharan West African Fulani, the North African Tuareg, and Early European Farmers, European agriculturalists, who are descendants of these Neolithic agriculturalists, share the lactase persistence variant –13910*T. While shared by Fulani and Tuareg herders, compared to the Tuareg variant, the Fulani variant of –13910*T has undergone a longer period of haplotype differentiation. The Fulani lactase persistence variant –13910*T may have spread, along with cattle pastoralism, between 9686 BP and 7534 BP, possibly around 8500 BP; corroborating this timeframe for the Fulani, by at least 7500 BP, there is evidence of herders engaging in the act of milking in the Central Sahara.West Africa
Archaic Human DNA
Archaic traits found in human fossils of West Africa (e.g., Iwo Eleru skull, Iho Eleru fossils, which dates to 13,000 BP) and Central Africa (e.g., Ishango fossils, which dates between 25,000 BP and 20,000 BP) may have developed as a result of admixture between archaic humans and modern humans or may be evidence of late-persisting Early modern human#Early Homo sapiens, early modern humans. While Denisovan and Neanderthal ancestry in Non-Africans (genetic lineage), non-Africans outside of Africa are more certain, archaic human ancestry in Africans is less certain and is too early to be established with certainty.Ancient DNA
As of 2017, human ancient DNA has not been found in the region of Western Africa. As of 2020, human ancient DNA has not been forthcoming in the region of Western Africa. In 4000 BP, there may have been a population that traversed from Africa (e.g., West Africa or West-Central Africa), through the Strait of Gibraltar, into the Iberian peninsula, where admixing between Africans and Iberians (e.g., of northern Portugal, of southern Spain) occurred. In Granada, a Muslim (Moors, Moor) of the Cordoba Caliphate, who was of haplogroups Haplogroup E-M2, E1b1a1 and Haplogroup H (mtDNA)#H1, H1+16189, as well as estimated to date between 900 CE and 1000 CE, and a Morisco, who was of Haplogroup L2 (mtDNA)#Haplogroup L2e, haplogroup L2e1, as well as estimated to date between 1500 CE and 1600 CE, were both found to be of Sub-Saharan West African (i.e., The Gambia#Ethnic groups, Gambian) and Iberian Peninsula, Iberian descent.Y-Chromosomal DNA
As a result of haplogroup D0, a basal branch of haplogroup DE, being found in three Nigerian men, it may be the case that haplogroup DE, as well as its sublineages D0 and E, originated in Africa. As of 19,000 years ago, Africans, bearing Haplogroup E-V38, haplogroup E1b1a-V38, likely traversed across the Sahara, from East Africa, east to West Africa, west. Haplogroup E-M2, E1b1a1-M2 likely originated in West Africa or Central Africa.Mitochondrial DNA
Between 75,000 BP and 60,000 BP, Africans bearing Haplogroup L3 (mtDNA), haplogroup L3 emerged in East Africa and eventually migrated into and became present in modern West Africans, Central Africans, and Non-Africans (genetic lineage), non-Africans. Amid the Holocene, including the Holocene Climate Optimum in 8000 BP, Africans bearing haplogroup L2 spread within West Africa and Africans bearing haplogroup L3 spread within East Africa. As the largest migration since the Out of Africa theory, Out of Africa migration, migration from Sub-Saharan Africa toward the North Africa occurred, by West Africans, Central Africans, and East Africans, resulting in migrations into Europe and Asia; consequently, Sub-Saharan African mitochondrial DNA was introduced into Europe and Asia. During the early period of the Holocene, 50% of Macro-haplogroup L (mtDNA), Sub-Saharan African mitochondrial DNA was introduced into North Africa by West Africa#Demographics and languages, West Africans and the other 50% was introduced by East Africa#Demographics, East Africans. During the modern period, a greater number of West Africans introduced Sub-Saharan African mitochondrial DNA into North Africa than East Africans. Between 15,000 BP and 7000 BP, 86% of Sub-Saharan African mitochondrial DNA was introduced into Southwest Asia by East Africa#Demographics, East Africans, largely in the region of Arabia, which constitute 50% of Sub-Saharan African mitochondrial DNA in modern Southwest Asia. In the modern period, 68% of Sub-Saharan African mitochondrial DNA was introduced by East Africans and 22% was introduced by West Africa#Demographics and languages, West Africans, which constitutes 50% of Sub-Saharan African mitochondrial DNA in modern Southwest Asia. During the early period of the Holocene, Sub-Saharan African mitochondrial DNA was introduced into Europe, mostly in Iberia. West Africa#Demographics and languages, West Africans probably migrated, across Sahelian Africa, North Africa, and the Strait of Gibraltar, into Europe, and introduced 63% of Sub-Saharan African mitochondrial DNA. During the modern period, West Africans introduced 75% of Sub-Saharan African mitochondrial DNA into Iberia and other parts of Europe, possibly by sea voyage. Around 18,000 BP, Mende people, along with The Gambia#Ethnic groups, Gambian peoples, grew in population size. In 15,000 BP, Niger-Congo speakers may have migrated from the Sahelian region of West Africa, along the Senegal River, and introduced Haplogroup L2 (mtDNA)#Haplogroup L2a1, L2a1 into North Africa, resulting in modern Demographics of Mauritania#Ethnic groups, Mauritanian peoples and Berbers of Tunisia inheriting it. Between 11,000 BP and 10,000 BP, Yoruba people and Esan people grew in population size. As early as 11,000 years ago, Sub-Saharan West Africans, bearing Macro-haplogroup L (mtDNA), macrohaplogroup L (e.g., Haplogroup L1 (mtDNA)#L1b, L1b1a11, L1b1a6a, L1b1a8, L1b1a9a1, Haplogroup L2 (mtDNA)#Haplogroup L2a1, L2a1k, Haplogroup L3 (mtDNA)#Subclade distribution, L3d1b1a), may have migrated through North Africa and into Europe, mostly into southern Europe (e.g., Iberia).Autosomal DNA
Between 2000 BP and 1500 BP, Nilo-Saharan-speakers may have migrated across the Sahel, from East Africa into West Africa, and admixed with Niger-Congo-speaking Berom people. A 2019 study on Fulani people by Fan et al., found that the Fulani show genetic affinity to isolated Afroasiatic languages, Afroasiatic-speaking groups in East Africa, Eastern Africa, specifically Omotic languages, Omotic-speakers such as the Aari people. While the Fulani have nearly exclusive indigenous African ancestry (defined by West and East African ancestry), they also show traces of West-Eurasian-like admixture, supporting an ancestral homeland somewhere in North or Eastern Africa, and westwards expansion during the Neolithic, possibly caused by the arrival and expansion of West-Eurasian-related groups.Medical DNA
=Sickle Cell
= Amid the Green Sahara, the mutation for sickle cell originated in the Sahara or in the Northwestern Congolian lowland forests, northwest forest region of western Central Africa (e.g., Cameroon) by at least 7,300 years ago, though possibly as early as 22,000 years ago. The ancestral sickle cell haplotype to modern haplotypes (e.g., Cameroon/Central African Republic and Benin/Senegal haplotypes) may have first arose in the ancestors of modern West Africans, bearing haplogroups Haplogroup E-M2#E1b1a1a1f, E1b1a1-L485 and Haplogroup E-M2#E1b1a1a1g, E1b1a1-U175 or their ancestral haplogroup E1b1a1-M4732. West Africans (e.g., Yoruba people, Yoruba and Esan people, Esan of Nigeria), bearing the Benin sickle cell haplotype, may have migrated through the Northeast Africa, northeastern region of Africa into the western region of Arabia. West Africans (e.g., Mende people, Mende of Sierra Leone), bearing the Senegal sickle cell haplotype, may have migrated into Mauritania (77% modern rate of occurrence) and Senegal (100%); they may also have migrated across the Sahara, into North Africa, and from North Africa, into Southern Europe, Turkey, and a region near northern Iraq and southern Turkey. Some may have migrated into and introduced the Senegal and Benin sickle cell haplotypes into Basra, Iraq, where both occur equally. West Africans, bearing the Benin sickle cell haplotype, may have migrated into the northern region of Iraq (69.5%), Jordan (80%), Lebanon (73%), Oman (52.1%), and Egypt (80.8%).=Schistosomes
= According to Steverding (2020), while not definite: Near the African Great Lakes, schistosomes (e.g., S. mansoni, S. haematobium) underwent evolution. Subsequently, there was an expansion alongside the Nile. From Egypt, the presence of schistosomes may have expanded, via migratory Yoruba people, into Western Africa. Thereafter, schistosomes may have expanded, via Bantu migration, migratory Bantu peoples, into the rest of Sub-Saharan Africa (e.g., Southern Africa, Central Africa).=Thalassemia
= Through pathways taken by Caravan (travellers), caravans, or via travel amid the Almoravid dynasty, Almovarid period, a population (e.g., Sub-Saharan West Africa#Demographics and languages, West Africans) may have introduced the –29 (A → G) Beta thalassemia, β-thalassemia mutation (found in notable amounts among African-Americans) into the North African region of Morocco.Central Africa
Archaic Human DNA
Archaic traits found in human fossils of West Africa (e.g., Iwo Eleru skull, Iho Eleru fossils, which dates to 13,000 BP) and Central Africa (e.g., Ishango fossils, which dates between 25,000 BP and 20,000 BP) may have developed as a result of admixture between archaic humans and modern humans or may be evidence of late-persisting Early modern human#Early Homo sapiens, early modern humans. While Denisovan and Neanderthal ancestry in Non-Africans (genetic lineage), non-Africans outside of Africa are more certain, archaic human ancestry in Africans is less certain and is too early to be established with certainty.Ancient DNA
In 4000 BP, there may have been a population that traversed from Africa (e.g., West Africa or West-Central Africa), through the Strait of Gibraltar, into the Iberian peninsula, where admixing between Africans and Iberians (e.g., of northern Portugal, of southern Spain) occurred.=Cameroon
= West African hunter-gatherers, in the region of western Central Africa (e.g., Shum Laka, Cameroon), particularly between 8000 BP and 3000 BP, were found to be related to modern Central African foragers, Central African hunter-gatherers (e.g., Baka people (Cameroon and Gabon), Baka, Bakola, Aka people, Biaka, Bedzan people, Bedzan).=Democratic Republic of Congo
= At Kindoki, in the Democratic Republic of Congo, there were three individuals, dated to the protohistoric period (230 BP, 150 BP, 230 BP); one carried haplogroups Haplogroup E-M2#E1b1a1a1d, E1b1a1a1d1a2 (E-CTS99, E-CTS99) and Haplogroup L1 (mtDNA)#L1c, L1c3a1b, another carried Haplogroup E-M96, haplogroup E (E-M96, E-PF1620), and the last carried haplogroups Haplogroup R1b#R1b (R-L278), R1b1 (R-P25 1, R-M415) and Haplogroup L0 (mtDNA)#Distribution, L0a1b1a1. At Ngongo Mbata, in the Democratic Republic of Congo, an individual, dated to the protohistoric period (220 BP), carried haplogroup Haplogroup L1 (mtDNA)#L1c, L1c3a. At Matangai Turu Northwest, in the Democratic Republic of Congo, an individual, dated to the Iron Age#Sub-Saharan Africa, Iron Age (750 BP), carried an undetermined haplogroup(s).Y-Chromosomal DNA
Haplogroup R1b#R1b1b (R-V88), Haplogroup R-V88 may have originated in western Central Africa (e.g., Equatorial Guinea), and, in the middle of the Holocene, arrived in North Africa through population migration.Mitochondrial DNA
In 150,000 BP, Africans (e.g., Central Africans, East Africans) bearing Haplogroup L1 (mtDNA), haplogroup L1 diverged. Between 75,000 BP and 60,000 BP, Africans bearing Haplogroup L3 (mtDNA), haplogroup L3 emerged in East Africa and eventually migrated into and became present in modern West Africans, Central Africans, and Non-Africans (genetic lineage), non-Africans. Amid the Holocene, including the Holocene Climate Optimum in 8000 BP, Africans bearing haplogroup L2 spread within West Africa and Africans bearing haplogroup L3 spread within East Africa. As the largest migration since the Out of Africa theory, Out of Africa migration, migration from Sub-Saharan Africa toward the North Africa occurred, by West Africans, Central Africans, and East Africans, resulting in migrations into Europe and Asia; consequently, Sub-Saharan African mitochondrial DNA was introduced into Europe and Asia. Mitochondrial Haplogroup L1 (mtDNA), haplogroup L1c is strongly associated with pygmies, especially with Mbenga people, Bambenga groups. L1c prevalence was variously reported as: 100% in Gyele language, Ba-Kola, 97% in Aka people, Aka (Ba-Benzélé), and 77% in Aka people, Biaka,Sarah A. Tishkoff et al. 2007History of Click-Speaking Populations of Africa Inferred from mtDNA and Y Chromosome Genetic Variation.
Molecular Biology and Evolution 2007 24(10):2180-2195 100% of the Bedzan people, Bedzan (Tikar), 97% and 100% in the Baka (Cameroon and Gabon), Baka people of Gabon and Cameroon, respectively, 97% in Bakoya (97%), and 82% in Bongo people (Gabon), Ba-Bongo. Mitochondrial haplogroups Haplogroup L2 (mtDNA), L2a and Haplogroup L0 (mtDNA), L0a are prevalent among the Bambuti.
Autosomal DNA
Genetically, African pygmies have some key difference between them and Bantu peoples.Medical DNA
Evidence suggests that, when compared to other Sub-Saharan African populations, African pygmy populations display unusually low levels of expression of the genes encoding for human growth hormone and Growth hormone receptor, its receptor associated with low Blood serum, serum levels of insulin-like growth factor-1 and short stature.Eastern Africa
Archaic Human DNA
While Denisovan and Neanderthal ancestry in Non-Africans (genetic lineage), non-Africans outside of Africa are more certain, archaic human ancestry in Africans is less certain and is too early to be established with certainty.Ancient DNA
=Ethiopia
= At Mota, Ethiopia, Mota, in Ethiopia, an individual, estimated to date to the 5th millennium BP, carried haplogroups Haplogroup E-P2, E1b1 and Haplogroup L3 (mtDNA)#Subclade distribution, L3x2a. The individual of Mota is genetically related to groups residing near the region of Mota, and in particular, are considerably genetically related to the Aari people, especially the blacksmith caste of that group.=Kenya
= At Jawuoyo Rockshelter, in Kisumu County, Kenya, a forager of the Later Stone Age carried haplogroups Haplogroup E-V68#E-V22, E1b1b1a1b2/E-V22 and Haplogroup L4 (mtDNA), L4b2a2c. At Ol Kalou, in Nyandarua County, Kenya, a pastoralist of the Pastoral Neolithic carried haplogroups Haplogroup E-Z827#E-M293, E1b1b1b2b2a1/E-M293 and Haplogroup L3 (mtDNA)#Subclade distribution, L3d1d. At Kokurmatakore, in Marsabit County, Kenya, a pastoralist of the Pastoral Iron Age#Sub-Saharan Africa, Iron Age carried haplogroups Haplogroup E-M35, E1b1b1/E-M35 and Haplogroup L3 (mtDNA)#Subclade distribution, L3a2a. At White Rock Point, in Homa Bay County, Kenya, there were two foragers of the Later Stone Age; one carried haplogroups Haplogroup BT, BT (xCT), likely Haplogroup B-M60, B, and Haplogroup L2 (mtDNA)#Haplogroup L2a, L2a4, and another probably carried Haplogroup L0 (mtDNA)#Distribution, haplogroup L0a2. At Nyarindi Rockshelter, in Kenya, there were two individuals, dated to the Later Stone Age (3500 BP); one carried Haplogroup L4 (mtDNA), haplogroup L4b2a and another carried Haplogroup E-M96, haplogroup E (E-M96, E-P162). At Lukenya Hill, in Kenya, there were two individuals, dated to the Pastoral Neolithic (3500 BP); one carried haplogroups Haplogroup E-Z827#E-V1515, E1b1b1b2b (E-M293, E-CTS10880) and Haplogroup L4 (mtDNA), L4b2a2b, and another carried Haplogroup L0 (mtDNA)#Distribution, haplogroup L0f1. At Hyrax Hill, in Kenya, an individual, dated to the Pastoral Neolithic (2300 BP), carried haplogroups Haplogroup E-Z827#E-V1515, E1b1b1b2b (E-M293, E-M293) and Haplogroup L5 (mtDNA), L5a1b. At Molo Cave, in Kenya, there were two individuals, dated to the Pastoral Neolithic (1500 BP); while one had haplogroups that went undetermined, another carried haplogroups Haplogroup E-Z827#E-V1515, E1b1b1b2b (E-M293, E-M293) and Haplogroup L3 (mtDNA)#Subclade distribution, L3h1a2a1. At Kakapel, in Kenya, there were three individuals, one dated to the Later Stone Age (3900 BP) and two dated to the Later Iron Age#Sub-Saharan Africa, Iron Age (300 BP, 900 BP); one carried haplogroups Haplogroup CT, CT (CT-M168, CT-M5695) and Haplogroup L3 (mtDNA)#Subclade distribution, L3i1, another carried Haplogroup L2 (mtDNA)#Haplogroup L2a1, haplogroup L2a1f, and the last carried Haplogroup L2 (mtDNA)#Haplogroup L2a, haplogroup L2a5. At Panga ya Saidi, in Kenya, an individual, estimated to date between 496 BP and 322 BP, carried haplogroups Haplogroup E-M215 (Y-DNA)#E-Z830 (E1b1b1b2), E1b1b1b2 and Haplogroup L4 (mtDNA), L4b2a2.Laikipia County At Kisima Farm/Porcupine Cave, in Laikipia County, Kenya, there were two pastoralists of the Pastoral Neolithic; one carried haplogroups Haplogroup E-Z827#E-M293, E1b1b1b2b2a1/E-M293 and Haplogroup M (mtDNA)#Haplogroup M1, M1a1, and another carried haplogroup Haplogroup M (mtDNA)#Haplogroup M1, M1a1f. At Kisima Farm/C4, in Laikipia County, Kenya, a pastoralist of the Pastoral Iron Age#Sub-Saharan Africa, Iron Age, carried haplogroups Haplogroup E-M75, E2 (xE2b)/E-M75 and Haplogroup L3 (mtDNA)#Subclade distribution, L3h1a1. At Laikipia District Burial, in Laikipia County, Kenya, a pastoralist of the Pastoral Iron Age#Sub-Saharan Africa, Iron Age carried haplogroup Haplogroup L0 (mtDNA)#Distribution, L0a1c1.
Nakuru County At Prettejohn's Gully, in Nakuru County, Kenya, there were two pastoralists of the early pastoral period; one carried haplogroups Haplogroup E-M75, E2 (xE2b)/E-M75 and Haplogroup K (mtDNA), K1a, and another carried haplogroup Haplogroup L3 (mtDNA)#Subclade distribution, L3f1b. At Cole's Burial, in Nakuru County, Kenya, a pastoralist of the Pastoral Neolithic carried haplogroups Haplogroup E-V68#Subclades of M78, E1b1b1a1a1b1/E-CTS3282 and Haplogroup L3 (mtDNA)#Subclade distribution, L3i2. At Rigo Cave, in Nakuru County, Kenya, there were three pastoralists of the Pastoral Neolithic/Elmenteitan, one carried haplogroups Haplogroup E-Z827#E-M293, E1b1b1b2b2a1/E-M293 and Haplogroup L3 (mtDNA)#Subclade distribution, L3f, another carried haplogroups Haplogroup E-Z827#E-V1515, E1b1b1b2b2/E-V1486, likely Haplogroup E-M215 (Y-DNA)#E-M293, E-M293, and probably Haplogroup M (mtDNA)#Haplogroup M1, M1a1b, and the last carried haplogroups Haplogroup E-M215 (Y-DNA)#E-M293, E1b1b1b2b2a1/E-M293 and Haplogroup L4 (mtDNA), L4b2a2c. At Naishi Rockshelter, in Nakuru County, Kenya, there two pastoralists of the Pastoral Neolithic; one carried haplogroups Haplogroup E-Z827#E-V1515, E1b1b1b2b/E-V1515, likely Haplogroup E-M215 (Y-DNA)#E-M293, E-M293, and Haplogroup L3 (mtDNA)#Subclade distribution, L3x1a, and another carried haplogroups Haplogroup A (Y-DNA)#Structure & subclade distribution, A1b (xA1b1b2a)/A-P108 and Haplogroup L0 (mtDNA)#Distribution, L0a2d. At Keringet Cave, in Nakuru County, Kenya, a pastoralist of the Pastoral Neolithic carried Haplogroup A (Y-DNA)#Structure & subclade distribution, haplogroups A1b1b2/A-L427 and Haplogroup L4 (mtDNA), L4b2a1, and another pastoralist of the Pastoral Neolithic/Elmenteitan carried Haplogroup K (mtDNA), haplogroup K1a. At Naivasha Burial Site, in Nakuru County, Kenya, there were five pastoralists of the Pastoral Neolithic; one carried Haplogroup L4 (mtDNA), haplogroup L4b2a2b, another carried haplogroups xBT, likely Haplogroup A (Y-DNA), A, and Haplogroup M (mtDNA)#Haplogroup M1, M1a1b, another carried haplogroups Haplogroup E-Z827#E-M293, E1b1b1b2b2a1/E-M293 and Haplogroup L3 (mtDNA)#Subclade distribution, L3h1a1, another carried haplogroups Haplogroup A (Y-DNA)#A1b1b2b (A-M13), A1b1b2b/A-M13 and Haplogroup L4 (mtDNA), L4a1, and the last carried haplogroups Haplogroup E-Z827#E-M293, E1b1b1b2b2a1/E-M293 and Haplogroup L3 (mtDNA)#Subclade distribution, L3x1a. At Njoro River Cave II, in Nakuru County, Kenya, a pastoralist of the Pastoral Neolithic carried haplogroup Haplogroup L3 (mtDNA)#Subclade distribution, L3h1a2a1. At Egerton Cave, in Nakuru County, Kenya, a pastoralist of the Pastoral Neolithic/Elmenteitan carried haplogroup Haplogroup L0 (mtDNA)#Distribution, L0a1d. At Ilkek Mounds, in Nakuru County, Kenya, a pastoralist of the Pastoral Iron Age#Sub-Saharan Africa, Iron Age carried haplogroups Haplogroup E-M75, E2 (xE2b)/E-M75 and Haplogroup L0 (mtDNA)#Distribution, L0f2a. At Deloraine Farm, in Nakuru County, Kenya, an iron metallurgist of the Iron Age#Sub-Saharan Africa, Iron Age carried haplogroups Haplogroup E-M2#E1b1a1a1a, E1b1a1a1a1a/E-M58 and Haplogroup L5 (mtDNA), L5b1.
Narok County At Kasiole 2, in Narok County, Kenya, a pastoralist of the Pastoral Iron Age#Sub-Saharan Africa, Iron Age carried haplogroups Haplogroup E-Z827#E-V1515, E1b1b1b2b/E-V1515, likely Haplogroup E-Z827#E-M293, E-M293, and Haplogroup L3 (mtDNA)#Subclade distribution, L3h1a2a1. At Emurua Ole Polos, in Narok County, Kenya, a pastoralist of the Pastoral Iron Age#Sub-Saharan Africa, Iron Age carried haplogroups Haplogroup E-Z827#E-M293, E1b1b1b2b2a1/E-M293 and Haplogroup L3 (mtDNA)#Subclade distribution, L3h1a2a1.
=Tanzania
= At Mlambalasi rockshelter, in Tanzania, an individual, dated between 20,345 BP and 17,025 BP, carried undetermined haplogroups. At Kisese II rockshelter, in Tanzania, an individual, dated between 7240 BP and 6985 BP, carried haplogroups Haplogroup B-M60#B-M112, B2b1a~ and Haplogroup L5 (mtDNA), L5b2. At Luxmanda, Tanzania, an individual, estimated to date between 3141 BP and 2890 BP, carried Haplogroup L2 (mtDNA)#Haplogroup L2a1, haplogroup L2a1. At Kuumbi Cave, Zanzibar, Kuumbi Cave, in Zanzibar, Tanzania, an individual, estimated to date between 1370 BP and 1303 BP, carried haplogroup Haplogroup L4 (mtDNA), L4b2a2c.Karatu District At Gishimangeda Cave, in Karatu District, Tanzania, there were eleven pastoralists of the Pastoral Neolithic; one carried haplogroups Haplogroup E-V68#Family tree, E1b1b1a1b2/E-V22 and Haplogroup HV (mtDNA)#Tree, HV1b1, another carried Haplogroup L0 (mtDNA)#Distribution, haplogroup L0a, another carried haplogroup Haplogroup L3 (mtDNA)#Subclade distribution, L3x1, another carried haplogroup Haplogroup L4 (mtDNA), L4b2a2b, another carried haplogroups Haplogroup E-Z827#E-M293, E1b1b1b2b2a1/E-M293 and Haplogroup L3 (mtDNA)#Subclade distribution, L3i2, another carried haplogroup Haplogroup L3 (mtDNA)#Subclade distribution, L3h1a2a1, another carried haplogroups Haplogroup E-Z827#E-V1515, E1b1b1b2b2/E-V1486, likely Haplogroup E-Z827#E-M293, E-M293 and Haplogroup L0 (mtDNA)#Distribution, L0f2a1, and another carried haplogroups Haplogroup E-Z827#E-V1515, E1b1b1b2b2/E-V1486, likely Haplogroup E-Z827#E-M293, E-M293, and Haplogroup T (mtDNA), T2+150; while most of the haplogroups among three pastoralists went undetermined, one was determined to carry haplogroup BT, likely Haplogroup B-M60, B.
Pemba Island At Makangale Cave, on Pemba Island, Tanzania, an individual, estimated to date between 1421 BP and 1307 BP, carried Haplogroup L0 (mtDNA)#Distribution, haplogroup L0a. At Makangale Cave, on Pemba Island, Tanzania, an individual, estimated to date between 639 BP and 544 BP, carried Haplogroup L2 (mtDNA)#Haplogroup L2a1a, haplogroup L2a1a2.
=Uganda
= At Munsa, in Uganda, an individual, dated to the Later Iron Age#Sub-Saharan Africa, Iron Age (500 BP), carried haplogroup Haplogroup L3 (mtDNA)#Subclade distribution, L3b1a1.Y-Chromosomal DNA
As of 19,000 years ago, Africans, bearing Haplogroup E-V38, haplogroup E1b1a-V38, likely traversed across the Sahara, from East Africa, east to West Africa, west. Before the Trans-Saharan slave trade#Trans-Saharan slave trade in the Middle Ages, slave Indian Ocean slave trade#Muslim Period, trade period, East Africans, who carried Haplogroup E-M2, haplogroup E1b1a-M2, expanded into Arabia, resulting in various rates of inheritance throughout Arabia (e.g., 2.8% Qatar, 3.2% Yemen, 5.5% United Arab Emirates, 7.4% Oman).Mitochondrial DNA
In 150,000 BP, Africans (e.g., Central Africans, East Africans) bearing Haplogroup L1 (mtDNA), haplogroup L1 diverged. In 130,000 BP, Africans bearing Haplogroup L5 (mtDNA), haplogroup L5 diverged in East Africa. Between 130,000 BP and 75,000 BP, behavioral modernity emerged among Southern Africans and long-term interactions between the regions of Southern Africa and Eastern Africa became established. Between 75,000 BP and 60,000 BP, Africans bearing Haplogroup L3 (mtDNA), haplogroup L3 emerged in East Africa and eventually migrated into and became present in modern West Africans, Central Africans, and Non-Africans (genetic lineage), non-Africans. Amid the Holocene, including the Holocene Climate Optimum in 8000 BP, Africans bearing haplogroup L2 spread within West Africa and Africans bearing haplogroup L3 spread within East Africa. As the largest migration since the Out of Africa theory, Out of Africa migration, migration from Sub-Saharan Africa toward the North Africa occurred, by West Africans, Central Africans, and East Africans, resulting in migrations into Europe and Asia; consequently, Sub-Saharan African mitochondrial DNA was introduced into Europe and Asia. During the early period of the Holocene, 50% of Macro-haplogroup L (mtDNA), Sub-Saharan African mitochondrial DNA was introduced into North Africa by West Africa#Demographics and languages, West Africans and the other 50% was introduced by East Africa#Demographics, East Africans. During the modern period, a greater number of West Africans introduced Sub-Saharan African mitochondrial DNA into North Africa than East Africans. Between 15,000 BP and 7000 BP, 86% of Sub-Saharan African mitochondrial DNA was introduced into Southwest Asia by East Africa#Demographics, East Africans, largely in the region of Arabia, which constitute 50% of Sub-Saharan African mitochondrial DNA in modern Southwest Asia. In the modern period, 68% of Sub-Saharan African mitochondrial DNA was introduced by East Africans and 22% was introduced by West Africans, which constitutes 50% of Sub-Saharan African mitochondrial DNA in modern Southwest Asia.Autosomal DNA
Across all areas of Madagascar, the average ancestry for the Malagasy people was found to be 4% West Eurasian, 37% Austronesian peoples, Austronesian, and 59% Bantu peoples, Bantu.Medical DNA
Southern Africa
Archaic Human DNA
While Denisovan and Neanderthal ancestry in Non-Africans (genetic lineage), non-Africans outside of Africa are more certain, archaic human ancestry in Africans is less certain and is too early to be established with certainty.Ancient DNA
Three Later Stone Age hunter-gatherers carried ancient DNA similar to Khoisan-speaking hunter-gatherers. Prior to the Bantu migration into the region, as evidenced by ancient DNA from Botswana, East African herders migrated into Southern Africa. Out of four Iron Age Bantu agriculturalists of West African origin, two earlier agriculturalists carried ancient DNA similar to Tsonga people, Tsonga and Venda people, Venda peoples and the two later agriculturalists carried ancient DNA similar to Nguni people; this indicates that there were various movements of peoples in the overall Bantu migration, which resulted in increased interaction and admixing between Bantu languages, Bantu-speaking peoples and Khoisan-speaking peoples.=Botswana
= At Nqoma, in Botswana, an individual, dated to the Early Iron Age#Sub-Saharan Africa, Iron Age (900 BP), carried Haplogroup L2 (mtDNA)#Haplogroup L2a1, haplogroup L2a1f. At Taukome, in Botswana, an individual, dated to the Early Iron Age#Sub-Saharan Africa, Iron Age (1100 BP), carried haplogroups Haplogroup E-M2#E1b1a1, E1b1a1 (E-M2, E-Z1123) and Haplogroup L0 (mtDNA)#Distribution, L0d3b1. At Xaro, in Botswana, there were two individuals, dated to the Early Iron Age#Sub-Saharan Africa, Iron Age (1400 BP); one carried haplogroups Haplogroup E-M2#E1b1a1a1c, E1b1a1a1c1a and Haplogroup L3 (mtDNA)#Subclade distribution, L3e1a2, and another carried haplogroups Haplogroup E-Z827#E-V1515, E1b1b1b2b (E-M293, E-CTS10880) and Haplogroup L0 (mtDNA)#Distribution, L0k1a2.=Malawi
=Fingira At Fingira rockshelter, in Malawi, an individual, dated between 6179 BP and 2341 BP, carried haplogroups Haplogroup B-M60#B-M182, B2 and Haplogroup L0 (mtDNA)#Distribution, L0d1. At Fingira, in Malawi, an individual, estimated to date between 6175 BP and 5913 BP, carried haplogroups Haplogroup BT, BT and Haplogroup L0 (mtDNA)#Distribution, L0d1b2b. At Fingira, in Malawi, an individual, estimated to date between 6177 BP and 5923 BP, carried haplogroups Haplogroup BT, BT and Haplogroup L0 (mtDNA)#Distribution, L0d1c. At Fingira, in Malawi, an individual, estimated to date between 2676 BP and 2330 BP, carried Haplogroup L0 (mtDNA)#Distribution, haplogroup L0f.
Chencherere At Chencherere, in Malawi, an individual, estimated to date between 5400 BP and 4800 BP, carried Haplogroup L0 (mtDNA)#Distribution, haplogroup L0k2. At Chencherere, in Malawi, an individual, estimated to date between 5293 BP and 4979 BP, carried Haplogroup L0 (mtDNA)#Distribution, haplogroup L0k1.
Hora At Hora 1 rockshelter, in Malawi, an individual, dated between 16,897 BP and 15,827 BP, carried haplogroups Haplogroup B-M60#B-M112, B2b and Haplogroup L5 (mtDNA), L5b. At Hora 1 rockshelter, in Malawi, an individual, dated between 16,424 BP and 14,029 BP, carried haplogroups Haplogroup B-M60#B-M112, B2b1a2~ and Haplogroup L0 (mtDNA)#Distribution, L0d3/L0d3b. At Hora, in Malawi, an individual, estimated to date between 10,000 BP and 5000 BP, carried haplogroups Haplogroup BT, BT and Haplogroup L0 (mtDNA)#Distribution, L0k2. At Hora, in Malawi, an individual, estimated to date between 8173 BP and 7957 BP, carried Haplogroup L0 (mtDNA)#Distribution, haplogroup L0a2.
=South Africa
= At Doonside, KwaZulu-Natal, Doonside, in South Africa, an individual, estimated to date between 2296 BP and 1910 BP, carried Haplogroup L0 (mtDNA)#Distribution, haplogroup L0d2. At Champagne Castle, in South Africa, an individual, estimated to date between 448 BP and 282 BP, carried Haplogroup L0 (mtDNA)#Distribution, haplogroup L0d2a1a. At Elands Bay Cave, Eland Cave, in South Africa, an individual, estimated to date between 533 BP and 453 BP, carried Haplogroup L3 (mtDNA)#Subclade distribution, haplogroup L3e3b1. At Mfongosi, in South Africa, an individual, estimated to date between 448 BP and 308 BP, carried Haplogroup L3 (mtDNA)#Subclade distribution, haplogroup L3e1b2. At Newcastle, KwaZulu-Natal, Newcastle, in South Africa, an individual, estimated to date between 508 BP and 327 BP, carried Haplogroup L3 (mtDNA)#Subclade distribution, haplogroup L3e2b1a2. At St Helena Bay, St. Helena, in South Africa, an individual, estimated to date between 2241 BP and 1965 BP, carried haplogroups Haplogroup A (Y-DNA)#A1b1b2a (A-M51), A1b1b2a and Haplogroup L0 (mtDNA)#Distribution, L0d2c1. At Faraoskop Rock Shelter, in South Africa, an individual, estimated to date between 2017 BP and 1748 BP, carried haplogroups Haplogroup A (Y-DNA)#A1b1b2a (A-M51), A1b1b2a and Haplogroup L0 (mtDNA)#Distribution, L0d1b2b1b. At Kasteelberg, in South Africa, an individual, estimated to date between 1282 BP and 1069 BP, carried haplogroup Haplogroup L0 (mtDNA)#Distribution, L0d1a1a. At Vaalkrans Shelter, in South Africa, an individual, estimated to date to 200 BP, is predominantly related to Khoisan speakers, partly related (15% - 32%) to East Africa#Demographics, East Africans, and carried haplogroups Haplogroup L0 (mtDNA)#Distribution, L0d3b1.Ballito Bay At Ballito Bay, South Africa, an individual, estimated to date between 1986 BP and 1831 BP, carried haplogroups Haplogroup A (Y-DNA)#Structure & subclade distribution, A1b1b2 and Haplogroup L0 (mtDNA)#Distribution, L0d2c1. At Ballito Bay, South Africa, an individual, estimated to date between 2149 BP and 1932 BP, carried haplogroups Haplogroup A (Y-DNA)#Structure & subclade distribution, A1b1b2 and Haplogroup L0 (mtDNA)#Distribution, L0d2a1.
=Zambia
= At Kalemba rockshelter, in Zambia, an individual, dated between 5285 BP and 4975 BP, carried Haplogroup L0 (mtDNA)#Distribution, haplogroup L0d1b2b.Y-Chromosomal DNA

 Various Y chromosome studies show that the San carry some of the most divergent (oldest) Human Y-chromosome DNA haplogroup, human Y-chromosome haplogroups. These haplogroups are specific sub-groups of haplogroups Haplogroup A (Y-DNA), A and Haplogroup B (Y-DNA), B, the two earliest branches on the human Y-chromosome phylogenetic tree, tree.
Various Y chromosome studies show that the San carry some of the most divergent (oldest) Human Y-chromosome DNA haplogroup, human Y-chromosome haplogroups. These haplogroups are specific sub-groups of haplogroups Haplogroup A (Y-DNA), A and Haplogroup B (Y-DNA), B, the two earliest branches on the human Y-chromosome phylogenetic tree, tree.
Mitochondrial DNA
In 200,000 BP, Africans (e.g., Khoisan of Southern Africa) bearing Haplogroup L0 (mtDNA), haplogroup L0 diverged from other Africans bearing haplogroup Haplogroup L1-6 (mtDNA), L1′6, which tend to be northward of Southern Africa. Between 130,000 BP and 75,000 BP, behavioral modernity emerged among Southern Africans and long-term interactions between the regions of Southern Africa and Eastern Africa became established. Mitochondrial DNA studies also provide evidence that the San carry high frequencies of the earliest Human mitochondrial DNA haplogroup, haplogroup branches in the human mitochondrial DNA tree. This DNA is inherited only from one's mother. The most divergent (oldest) mitochondrial haplogroup, Haplogroup L0 (mtDNA), L0d, has been identified at its highest frequencies in the southern African San groups.Autosomal DNA
In a study published in March 2011, Brenna Henn and colleagues found that the ǂKhomani San, as well as the Sandawe people, Sandawe and Hadza peoples of Tanzania, were the most genetically diverse of any living humans studied. This high degree of genetic diversity hints at the origin of anatomically modern humans.Medical DNA
Among the ancient DNA from three hunter-gatherers sharing genetic similarity with San people and four Iron Age agriculturalists, their SNPs indicated that they bore variants for resistance against sleeping sickness and Plasmodium vivax. In particular, two out of the four Iron Age agriculturalists bore variants for resistance against sleeping sickness and three out of the four Iron Age agriculturalists bore Human genetic resistance to malaria#Duffy antigen receptor negativity, Duffy negative variants for resistance against malaria. In contrast to the Iron Age agriculturalists, from among the San-related hunter-gatherers, a six-year-old boy may have died from schistosomiasis. In Botswana, a man, who dates to 1400 BP, may have also carried the Duffy negative variant for resistance against malaria.Recent African origin of modern humans
Between 500,000 BP and 300,000 BP, anatomically modern humans may have emerged in Africa. As Africans (e.g., Y-Chromosomal Adam, Mitochondrial Eve) have migrated from their places of origin in Africa to other locations in Africa, and as the time of divergence for East African, Central African, and West African lineages are similar to the time of divergence for the Southern African lineage, there is insufficient evidence to identify a specific region for the origin of humans in Africa. In 100,000 BP, anatomically modern humans migrated from Africa into Eurasia. Subsequently, tens of thousands of years after, the ancestors of all present-day Eurasians migrated from Africa into Eurasia and eventually became admixed with Denisovans and Neanderthals. Archaeological and fossil evidence provide support for the recent African origin of modern humans, African origin of homo sapiens and behavioral modernity. Models reflecting a pan-African origin (multiple locations of origin within Africa) and evolution of modern humans have been developed. As the idea of "modern" has become increasingly problematized, research has "begun to disentangle what is meant by "modern" genetic ancestry, skeletal morphology, and behavior, recognizing these are unlikely to form a single package."See also
* Genetic history of the African diasporaNotes
References
{{Human genetics Genetic genealogy History of Africa Modern human genetic history