DNA-encoded chemical library on:
[Wikipedia]
[Google]
[Amazon]
DNA-encoded chemical libraries (DECL) is a technology for the
 The concept of DNA-encoding was first described in a theoretical paper by
The concept of DNA-encoding was first described in a theoretical paper by
 A promising strategy for the construction of DNA-encoded libraries is represented by the use of multifunctional building blocks
A promising strategy for the construction of DNA-encoded libraries is represented by the use of multifunctional building blocks
 Encoded Self-Assembling Chemical (ESAC) libraries rely on the principle that two sublibraries of a size of x members (e.g. 103) containing a constant complementary hybridization domain can yield a combinatorial DNA-duplex library after hybridization with a complexity of x2 uniformly represented library members (e.g. 106). Each sub-library member would consist of an
Encoded Self-Assembling Chemical (ESAC) libraries rely on the principle that two sublibraries of a size of x members (e.g. 103) containing a constant complementary hybridization domain can yield a combinatorial DNA-duplex library after hybridization with a complexity of x2 uniformly represented library members (e.g. 106). Each sub-library member would consist of an
 In 2001 David Liu and co-workers showed that complementary DNA
In 2001 David Liu and co-workers showed that complementary DNA
 The center of the DNA junction constitutes a volume on the order of a
The center of the DNA junction constitutes a volume on the order of a
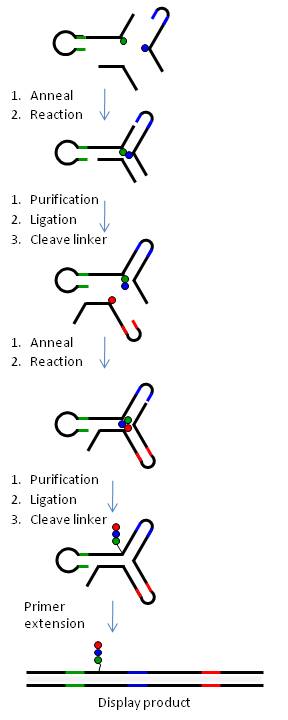 In summary, chemical building-blocks (BB) are attached via cleavable or non-cleavable linkers to three types of bispecific DNA oligonucleotides (oligo-BBs) representing each arm of the yR. To facilitate synthesis in a combinatorial manner, the oligo-BBs are designed such that the DNA contains (a) the code for an attached BB at the distal end of the oligo (colored lines) and (b) areas of constant DNA sequence (black lines) to bring about the self-assembly of the DNA into a 3-way junction (independently of the BB) and the subsequent chemical reaction. Chemical reactions are performed via a stepwise procedure and after each step the DNA is ligated and the product purified by polyacrylamide gel electrophoresis. Cleavable linkers (BB-DNA) are used for all but one position yielding a library of small molecules with a single covalent link to the DNA code. Table 1 outlines how libraries of different sizes can be generated using yR technology.
In summary, chemical building-blocks (BB) are attached via cleavable or non-cleavable linkers to three types of bispecific DNA oligonucleotides (oligo-BBs) representing each arm of the yR. To facilitate synthesis in a combinatorial manner, the oligo-BBs are designed such that the DNA contains (a) the code for an attached BB at the distal end of the oligo (colored lines) and (b) areas of constant DNA sequence (black lines) to bring about the self-assembly of the DNA into a 3-way junction (independently of the BB) and the subsequent chemical reaction. Chemical reactions are performed via a stepwise procedure and after each step the DNA is ligated and the product purified by polyacrylamide gel electrophoresis. Cleavable linkers (BB-DNA) are used for all but one position yielding a library of small molecules with a single covalent link to the DNA code. Table 1 outlines how libraries of different sizes can be generated using yR technology.
 The yR design approach provides an unvarying reaction site with regard to both (a) distance between reactants and (b) sequence environment surrounding the reaction site. Furthermore, the intimate connection between the code and the BB on the oligo-BB moieties which are mixed combinatorially in a single pot confers a high fidelity to the encoding of the library. The code of the synthesized products, furthermore, is not preset, but rather is assembled combinatorially and synthesized in synchronicity with the innate product.
The yR design approach provides an unvarying reaction site with regard to both (a) distance between reactants and (b) sequence environment surrounding the reaction site. Furthermore, the intimate connection between the code and the BB on the oligo-BB moieties which are mixed combinatorially in a single pot confers a high fidelity to the encoding of the library. The code of the synthesized products, furthermore, is not preset, but rather is assembled combinatorially and synthesized in synchronicity with the innate product.
 This intracellular screening technique allows for the discovery of ligands that engage targets in their native conformation and cellular environment, enabling screening of targets that are difficult to express or purify, and potentially improving the physiological relevance of identified compounds.
This intracellular screening technique allows for the discovery of ligands that engage targets in their native conformation and cellular environment, enabling screening of targets that are difficult to express or purify, and potentially improving the physiological relevance of identified compounds.
synthesis
Synthesis or synthesize may refer to:
Science Chemistry and biochemistry
*Chemical synthesis, the execution of chemical reactions to form a more complex molecule from chemical precursors
**Organic synthesis, the chemical synthesis of organi ...
and screening on an unprecedented scale of collections of small molecule
In molecular biology and pharmacology, a small molecule or micromolecule is a low molecular weight (≤ 1000 daltons) organic compound that may regulate a biological process, with a size on the order of 1 nm. Many drugs are small molecules; ...
compounds. DECL is used in medicinal chemistry
Medicinal or pharmaceutical chemistry is a scientific discipline at the intersection of chemistry and pharmacy involved with drug design, designing and developing pharmaceutical medication, drugs. Medicinal chemistry involves the identification, ...
to bridge the fields of combinatorial chemistry
Combinatorial chemistry comprises chemical synthesis, chemical synthetic methods that make it possible to prepare a large number (tens to thousands or even millions) of compounds in a single process. These compound library, compound libraries can b ...
and molecular biology
Molecular biology is a branch of biology that seeks to understand the molecule, molecular basis of biological activity in and between Cell (biology), cells, including biomolecule, biomolecular synthesis, modification, mechanisms, and interactio ...
. The aim of DECL technology is to accelerate the drug discovery
In the fields of medicine, biotechnology, and pharmacology, drug discovery is the process by which new candidate medications are discovered.
Historically, drugs were discovered by identifying the active ingredient from traditional remedies or ...
process and in particular early phase discovery activities such as target validation and hit identification.
DECL technology involves the conjugation
Conjugation or conjugate may refer to:
Linguistics
*Grammatical conjugation, the modification of a verb from its basic form
*Emotive conjugation or Russell's conjugation, the use of loaded language
Mathematics
*Complex conjugation, the change o ...
of chemical compounds or building blocks to short DNA
Deoxyribonucleic acid (; DNA) is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of al ...
fragments that serve as identification bar codes and in some cases also direct and control the chemical synthesis. The technique enables the mass creation and interrogation of libraries via affinity selection, typically on an immobilized protein target. A homogeneous method for screening DNA-encoded libraries (DELs) has recently been developed which uses water-in-oil emulsion technology to isolate, count and identify individual ligand-target complexes in a single-tube approach. In contrast to conventional screening procedures such as high-throughput screening
High-throughput screening (HTS) is a method for scientific discovery especially used in drug discovery and relevant to the fields of biology, materials science and chemistry. Using robotics, data processing/control software, liquid handling device ...
, biochemical assays are not required for binder identification, in principle allowing the isolation of binders to a wide range of proteins historically difficult to tackle with conventional screening technologies. So, in addition to the general discovery of target specific molecular compounds, the availability of binders to pharmacologically important, but so-far “undruggable” target proteins opens new possibilities to develop novel drugs for diseases that could not be treated so far. In eliminating the requirement to initially assess the activity of hits it is hoped and expected that many of the high affinity binders identified will be shown to be active in independent analysis of selected hits, therefore offering an efficient method to identify high quality hits and pharmaceutical leads.
DNA-encoded chemical libraries and display technologies
Until recently, the application ofmolecular evolution
Molecular evolution describes how Heredity, inherited DNA and/or RNA change over evolutionary time, and the consequences of this for proteins and other components of Cell (biology), cells and organisms. Molecular evolution is the basis of phylogen ...
in the laboratory had been limited to display technologies involving biological molecules, where small molecules lead discovery was considered beyond this biological approach. DELs have opened the field of display technology to include non-natural compounds such as small molecules, extending the application of molecular evolution and natural selection to the identification of small molecule compounds of desired activity and function.
DNA encoded chemical libraries bear resemblance to biological display technologies such as antibody phage display technology, yeast display, mRNA display
mRNA display is a display technique used for ''in vitro'' protein, and/or peptide evolution to create molecules that can bind to a desired target. The process results in translated peptides or proteins that are associated with their mRNA progenitor ...
and aptamer SELEX. In antibody phage display, antibodies are physically linked to phage particles that bear the gene coding for the attached antibody, which is equivalent to a physical linkage of a “phenotype
In genetics, the phenotype () is the set of observable characteristics or traits of an organism. The term covers the organism's morphology (physical form and structure), its developmental processes, its biochemical and physiological propert ...
” (the protein) and a “genotype
The genotype of an organism is its complete set of genetic material. Genotype can also be used to refer to the alleles or variants an individual carries in a particular gene or genetic location. The number of alleles an individual can have in a ...
” (the gene encoding for the protein ). Phage-displayed antibodies can be isolated from large antibody libraries by mimicking molecular evolution
Molecular evolution describes how Heredity, inherited DNA and/or RNA change over evolutionary time, and the consequences of this for proteins and other components of Cell (biology), cells and organisms. Molecular evolution is the basis of phylogen ...
: through rounds of selection (on an immobilized protein target), amplification and translation.
In DELs the linkage of a small molecule to an identifier DNA code allows the facile identification of binding molecules. DELs are subjected to affinity selection procedures on an immobilized target protein of choice, after which non-binders are removed by washing steps, and binders can subsequently be amplified by polymerase chain reaction (PCR) and identified by virtue of their DNA code (e.g.by DNA sequencing). In evolution-based DEL technologies hits can be further enriched by performing rounds of selection, PCR amplification and translation in analogy to biological display systems such as antibody phage display. This makes it possible to work with much larger libraries.
History
“Synthesize a multi-component mixture of compounds in a single process and screen it also a single process”. This is the principle of combinatorial chemistry invented by Prof. Furka Á. (Eötvös Loránd University Budapest Hungary) in 1982, and described it including the method of synthesis of combinatorial libraries and that of a deconvolution strategy in a document notarized in the same year. Motivations that led to the invention had been published in 2002. DNA encoded chemical libraries (DECLs) are synthesized by the combinatorial chemistry principle and it clearly agrees with their application.Sydney Brenner
Sydney Brenner (13 January 1927 – 5 April 2019) was a South African biologist. In 2002, he shared the Nobel Prize in Physiology or Medicine with H. Robert Horvitz and Sir John E. Sulston. Brenner made significant contributions to wo ...
and Richard Lerner
Richard Alan Lerner (August 26, 1938 – December 2, 2021) was an American research chemist. He was best known for his work on catalytic antibodies and combinatorial antibody libraries. Lerner served as President of The Scripps Research Inst ...
in 1992 in which was proposed to link each molecule of a chemically synthesized entity to a particular oligonucleotide
Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, Recombinant DNA, research, and Forensic DNA, forensics. Commonly made in the laboratory by Oligonucleotide synthesis, solid-phase ...
sequence constructed in parallel and to use this encoding genetic tag to identify and enrich active compounds. In 1993 the first practical implementation of this approach was presented by J Nielsen, S. Brenner and K. Janda and similarly by the group of M.A. Gallop. Brenner and Janda suggested to generate individual encoded library members by an alternating parallel combinatorial synthesis
Combinatorial chemistry comprises chemical synthetic methods that make it possible to prepare a large number (tens to thousands or even millions) of compounds in a single process. These compound libraries can be made as mixtures, sets of individu ...
of the heteropolymeric chemical compound and the appropriate oligonucleotide sequence on the same bead in a “split-&-pool”-based fashion (see below).
Since unprotected DNA is restricted to a narrow window of conventional reaction conditions, until the end of the 1990s a number of alternative encoding strategies were envisaged (i.e. MS-based compound tagging, peptide
Peptides are short chains of amino acids linked by peptide bonds. A polypeptide is a longer, continuous, unbranched peptide chain. Polypeptides that have a molecular mass of 10,000 Da or more are called proteins. Chains of fewer than twenty am ...
encoding, haloaromatic
In organic chemistry, an aryl halide (also known as a haloarene) is an aromatic compound in which one or more hydrogen atoms directly bonded to an aromatic ring are replaced by a halide ion (such as fluorine F''−'', chlorine Cl−1,−3,−5, bro ...
tagging, encoding by secondary amine
In chemistry, amines (, ) are organic compounds that contain carbon-nitrogen bonds. Amines are formed when one or more hydrogen atoms in ammonia are replaced by alkyl or aryl groups. The nitrogen atom in an amine possesses a lone pair of elec ...
s, semiconductor
A semiconductor is a material with electrical conductivity between that of a conductor and an insulator. Its conductivity can be modified by adding impurities (" doping") to its crystal structure. When two regions with different doping level ...
devices.), mainly to avoid inconvenient solid phase DNA synthesis and to create easily screenable combinatorial libraries in high-throughput fashion. However, the selective amplificability of DNA greatly facilitates library screening and it becomes indispensable for the encoding of organic compounds libraries of this unprecedented size. Consequently, at the beginning of the 2000s DNA-combinatorial chemistry experienced a revival.
The beginning of the millennium saw the introduction of several independent developments in DEL technology. These technologies can be classified under two general categories: non-evolution-based and evolution-based DEL technologies capable of molecular evolution
Molecular evolution describes how Heredity, inherited DNA and/or RNA change over evolutionary time, and the consequences of this for proteins and other components of Cell (biology), cells and organisms. Molecular evolution is the basis of phylogen ...
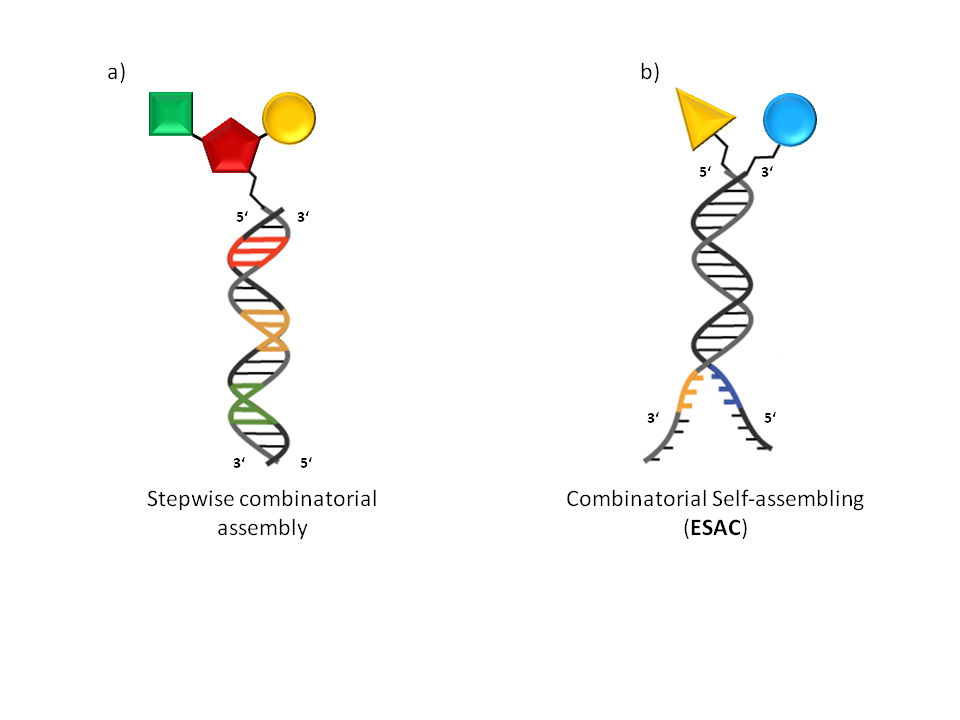
. The first category benefits from the ability to use off the shelf reagents and therefore enables rather straightforward library generation. Hits can be identified by DNA sequencing, however DNA translation and therefore molecular evolution is not feasible by these methods. The split and pool approaches developed by researchers at Praecis Pharmaceuticals (now owned by GlaxoSmithKline), Nuevolution (Copenhagen, Denmark) and encoded self- assembled chemical (ESAC) technology developed in the laboratory of Prof D. Neri (Institute of Pharmaceutical Science, Zurich, Switzerland) fall under this category. ESAC technology sets itself apart being a combinatorial self-assembling approach which resembles fragment based hit discovery (Fig 1b). Here DNA annealing enables discrete building block combinations to be sampled, but no chemical reaction takes place between them.
Examples of evolution-based DEL technologies are DNA-routing developed by Prof. D.R. Halpin and Prof. P.B. Harbury (Stanford University, Stanford, CA), DNA-templated synthesis developed by Prof. D. Liu (Harvard University, Cambridge, MA) and commercialized by Ensemble Therapeutics (Cambridge, MA) and YoctoReactor technology. developed and commercialized by Vipergen (Copenhagen, Denmark). These technologies are described in further detail below. DNA-templated synthesis and YoctoReactor technology require the prior conjugation of chemical building blocks (BB) to a DNA oligonucleotide tag before library assembly, therefore more upfront work is required before library assembly. Furthermore, the DNA tagged BBs enable the generation of a genetic code for synthesized compounds and artificial translation of the genetic code is possible: That is the BB's can be recalled by the PCR-amplified genetic code, and the library compounds can be regenerated. This, in turn, enables the principle of Darwinian natural selection and evolution to be applied to small molecule selection in direct analogy to biological display systems; through rounds of selection, amplification and translation.
Non-evolution based technologies
Combinatorial libraries
Combinatorial libraries are special multi-component compound mixtures that are synthesized in a single stepwise process. They differ from collection of individual compounds as well as from a series of compounds prepared by parallel synthesis. Combinatorial libraries have important features. ″ Mixtures are used in their synthesis. The use of mixtures ensures the very high efficiency of the process. Both reactants could be mixtures but for practical reasons the split-mix procedure is used: one mixture is divided into portions that are coupled with the BBs.Á. Furka, F. Sebestyén, M. Asgedom, G. Dibó, Cornucopia of peptides by synthesis In Highlights of Modern Biochemistry, Proceedings of the 14th International Congress of Biochemistry, VSP. Utrecht, The Netherlands, 1988, Vol. 5, p 47.Furka Á, Sebestyén F, Asgedom M, Dibó G ( 1991) General method for rapid synthesis of multicomponent peptide mixtures. Int J Peptide Protein Res 37; 487-93. The mixtures are so important that there is no combinatorial library without using a mixture in the synthesis, and if a mixture is used in a process inevitably combinatorial library forms. ″ Components of the libraries need to be present in nearly equal molar quantities. In order to achieve this as closely as possible the mixtures are divided into equal portions and after pooling a thorough mixing is needed. ″ Since the structure of components is unknown deconvolution methods need to be used in screening. For this reason, encoding methods had been developed. Coding molecules are attached to the beads of the solid support that record the coupled BBs and their sequence. One of these methods is encoding by DNA oligomers. ″ It is a remarkable feature of combinatorial libraries that the whole compound mixture can be screened in a single process. Since both the synthesis and screening are very efficient procedures the use of combinatorial libraries in pharmaceutical research leads to enormous savings. In solid phase combinatorial synthesis only a single compound forms in each bead. For this reason, the number of components in the library can't exceed the number of beads of the solid support. This means that the number of components in such libraries is limited. This restraint was eliminated by Harbury and Halpin. In their synthesis of DELs, the solid support is omitted and BBs are attached directly to the encoding DNA oligomers. This new approach helps to increase practically unlimitedly the number of components of DNA encoded combinatorial libraries (DECLs).Split-&-Pool DNA Encoding
In order to applycombinatorial chemistry
Combinatorial chemistry comprises chemical synthesis, chemical synthetic methods that make it possible to prepare a large number (tens to thousands or even millions) of compounds in a single process. These compound library, compound libraries can b ...
for the synthesis of DNA-encoded chemical libraries, a Split-&-Pool approach was pursued. Initially a set of unique DNA-oligonucleotides
Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, research, and forensics. Commonly made in the laboratory by solid-phase chemical synthesis, these small fragments of nucleic aci ...
(n) each containing a specific coding sequence is chemically conjugated to a corresponding set of small organic molecules. Consequently, the oligonucleotide
Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, Recombinant DNA, research, and Forensic DNA, forensics. Commonly made in the laboratory by Oligonucleotide synthesis, solid-phase ...
-conjugate compounds are mixed ("Pool") and divided ("Split") into a number of groups (m). In appropriate conditions a second set of building blocks (m) are coupled to the first one and a further oligonucleotide
Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, Recombinant DNA, research, and Forensic DNA, forensics. Commonly made in the laboratory by Oligonucleotide synthesis, solid-phase ...
which is coding for the second modification is enzymatically introduced before mixing again. This “split-&-pool” steps can be iterated a number of times (r) increasing at each round the library size in a combinatorial manner (i.e. (n x m)r). Alternatively, peptide nucleic acids have been used to encode libraries prepared by "split-&-pool" method. A benefit of PNA-encoding is that the chemistry can be performed by standard SPPS.
Stepwise coupling of coding DNA fragments to nascent organic molecules
 A promising strategy for the construction of DNA-encoded libraries is represented by the use of multifunctional building blocks
A promising strategy for the construction of DNA-encoded libraries is represented by the use of multifunctional building blocks covalent
A covalent bond is a chemical bond that involves the sharing of electrons to form electron pairs between atoms. These electron pairs are known as shared pairs or bonding pairs. The stable balance of attractive and repulsive forces between atom ...
ly conjugated to an oligonucleotide
Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, Recombinant DNA, research, and Forensic DNA, forensics. Commonly made in the laboratory by Oligonucleotide synthesis, solid-phase ...
serving as a “core structure” for library synthesis. In a ‘pool-and-split’ fashion a set of multifunctional scaffolds undergo orthogonal reactions with series of suitable reactive partners. Following each reaction step, the identity of the modification is encoded by an enzymatic addition of DNA segment to the original DNA “core structure”. The use of ''N''-protected amino acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although over 500 amino acids exist in nature, by far the most important are the 22 α-amino acids incorporated into proteins. Only these 22 a ...
s covalently attached to a DNA fragment allow, after a suitable deprotection step, a further amide bond
In organic chemistry, an amide, also known as an organic amide or a carboxamide, is a compound with the general formula , where R, R', and R″ represent any group, typically organyl groups or hydrogen atoms. The amide group is called a p ...
formation with a series of carboxylic acid
In organic chemistry, a carboxylic acid is an organic acid that contains a carboxyl group () attached to an Substituent, R-group. The general formula of a carboxylic acid is often written as or , sometimes as with R referring to an organyl ...
s or a reductive amination
Reductive amination (also known as reductive alkylation) is a form of amination that converts a carbonyl group to an amine via an intermediate imine. The carbonyl group is most commonly a ketone or an aldehyde. It is a common method to make amine ...
with aldehydes
In organic chemistry, an aldehyde () (lat. ''al''cohol ''dehyd''rogenatum, dehydrogenated alcohol) is an organic compound containing a functional group with the structure . The functional group itself (without the "R" side chain) can be referred ...
. Similarly, diene
In organic chemistry, a diene ( ); also diolefin, ) or alkadiene) is a covalent compound that contains two double bonds, usually among carbon atoms. They thus contain two alk''ene'' units, with the standard prefix ''di'' of systematic nome ...
carboxylic acids used as scaffolds for library construction at the 5’-end of amino modified oligonucleotide
Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, Recombinant DNA, research, and Forensic DNA, forensics. Commonly made in the laboratory by Oligonucleotide synthesis, solid-phase ...
, could be subjected to a Diels-Alder reaction with a variety of maleimide
Maleimide is a chemical compound with the formula H2C2(CO)2NH (see diagram). This unsaturated imide is an important building block in organic synthesis. The name is a contraction of maleic acid and imide, the -C(O)NHC(O)- functional group. Malei ...
derivatives. After completion of the desired reaction step, the identity of the chemical moiety added to the oligonucleotide
Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, Recombinant DNA, research, and Forensic DNA, forensics. Commonly made in the laboratory by Oligonucleotide synthesis, solid-phase ...
is established by the annealing of a partially complementary oligonucleotide
Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, Recombinant DNA, research, and Forensic DNA, forensics. Commonly made in the laboratory by Oligonucleotide synthesis, solid-phase ...
and by a subsequent Klenow fill-in DNA-polymerization, yielding a double stranded DNA fragment. The synthetic and encoding strategies described above enable the facile construction of DNA-encoded libraries of a size up to 104 member compounds carrying two sets of “building blocks”. However the stepwise addition of at least three independent sets of chemical moieties to a tri-functional core building block for the construction and encoding of a very large DNA-encoded library (comprising up to 106 compounds) can also be envisaged.(Fig.2)
Combinatorial self-assembling
Encoded self-assembling chemical libraries
 Encoded Self-Assembling Chemical (ESAC) libraries rely on the principle that two sublibraries of a size of x members (e.g. 103) containing a constant complementary hybridization domain can yield a combinatorial DNA-duplex library after hybridization with a complexity of x2 uniformly represented library members (e.g. 106). Each sub-library member would consist of an
Encoded Self-Assembling Chemical (ESAC) libraries rely on the principle that two sublibraries of a size of x members (e.g. 103) containing a constant complementary hybridization domain can yield a combinatorial DNA-duplex library after hybridization with a complexity of x2 uniformly represented library members (e.g. 106). Each sub-library member would consist of an oligonucleotide
Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, Recombinant DNA, research, and Forensic DNA, forensics. Commonly made in the laboratory by Oligonucleotide synthesis, solid-phase ...
containing a variable, coding region flanked by a constant DNA sequence, carrying a suitable chemical modification at the oligonucleotide extremity. The ESAC sublibraries can be used in at least four different embodiments.
* A sub-library can be paired with a complementary oligonucleotide and used as a DNA encoded library displaying a single covalently linked compound for affinity-based selection experiments.
* A sub-library can be paired with an oligonucleotide displaying a known binder to the target, thus enabling affinity maturation strategies.
* Two individual sublibraries can be assembled combinatorially and used for the ''de novo'' identification of bidentate binding molecules.
* Three different sublibraries can be assembled to form a combinatorial triplex library.
Preferential binders isolated from an affinity-based selection can be PCR-amplified and decoded on complementary oligonucleotide
Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, Recombinant DNA, research, and Forensic DNA, forensics. Commonly made in the laboratory by Oligonucleotide synthesis, solid-phase ...
microarrays
A microarray is a multiplex lab-on-a-chip. Its purpose is to simultaneously detect the expression of thousands of biological interactions. It is a two-dimensional array on a solid substrate—usually a glass slide or silicon thin-film cell� ...
or by concatenation of the codes, subcloning
In molecular biology, subcloning is a technique used to move a particular DNA sequence from a ''parent vector'' to a ''destination vector''.
Subcloning is not to be confused with molecular cloning, a related technique.
Procedure
Restriction ...
and sequencing
In genetics and biochemistry, sequencing means to determine the primary structure (sometimes incorrectly called the primary sequence) of an unbranched biopolymer. Sequencing results in a symbolic linear depiction known as a sequence which succ ...
. The individual building blocks can eventually be conjugated using suitable linkers to yield a drug-like high-affinity compound. The characteristics of the linker (e.g. length, flexibility, geometry, chemical nature and solubility) influence the binding affinity
In biochemistry and pharmacology, a ligand is a substance that forms a complex with a biomolecule to serve a biological purpose. The etymology stems from Latin ''ligare'', which means 'to bind'. In protein-ligand binding, the ligand is usuall ...
and the chemical properties of the resulting binder.(Fig.3)
Bio-panning experiments on HSA of a 600-member ESAC library allowed the isolation of the 4-(''p''-iodophenyl)butanoic moiety. The compound represents the core structure of a series of portable albumin
Albumin is a family of globular proteins, the most common of which are the serum albumins. All of the proteins of the albumin family are water- soluble, moderately soluble in concentrated salt solutions, and experience heat denaturation. Alb ...
binding molecules and of Albufluor a recently developed fluorescein
Fluorescein is an organic compound and dye based on the xanthene tricyclic structural motif, formally belonging to Triarylmethane dye, triarylmethine dyes family. It is available as a dark orange/red powder slightly soluble in water and alcohol. ...
angiographic
Angiography or arteriography is a medical imaging technique used to visualize the inside, or lumen, of blood vessels and organs of the body, with particular interest in the arteries, veins, and the heart chambers. Modern angiography is perform ...
contrast agent
A contrast agent (or contrast medium) is a substance used to increase the contrast of structures or fluids within the body in medical imaging. Contrast agents absorb or alter external electromagnetism or ultrasound, which is different from radiop ...
currently under clinical evaluation.
ESAC technology has been used for the isolation of potent inhibitors of bovine trypsin
Trypsin is an enzyme in the first section of the small intestine that starts the digestion of protein molecules by cutting long chains of amino acids into smaller pieces. It is a serine protease from the PA clan superfamily, found in the dig ...
and for the identification of novel inhibitors of stromelysin-1 ( MMP-3), a matrix metalloproteinase involved in both physiological and pathological tissue remodeling processes, as well as in disease processes, such as arthritis
Arthritis is a general medical term used to describe a disorder that affects joints. Symptoms generally include joint pain and stiffness. Other symptoms may include redness, warmth, Joint effusion, swelling, and decreased range of motion of ...
and metastasis
Metastasis is a pathogenic agent's spreading from an initial or primary site to a different or secondary site within the host's body; the term is typically used when referring to metastasis by a cancerous tumor. The newly pathological sites, ...
.
Evolution-based technologies
DNA-routing
In 2004, D.R. Halpin and P.B. Harbury presented a novel intriguing method for the construction of DNA-encoded libraries. For the first time the DNA-conjugated templates served for both encoding and programming the infrastructure of the “split-&-pool” synthesis of the library components. The design of Halpin and Harbury enabled alternating rounds of selection,PCR amplification
The polymerase chain reaction (PCR) is a method widely used to make millions to billions of copies of a specific DNA sample rapidly, allowing scientists to amplify a very small sample of DNA (or a part of it) sufficiently to enable detailed st ...
and diversification with small organic molecules, in complete analogy to phage display
Phage display is a laboratory technique for the study of protein–protein, protein–peptide, and protein–DNA interactions that uses bacteriophages (viruses that infect bacteria) to connect proteins with the genetic information that encodes ...
technology. The DNA-routing machinery consists of a series of connected columns bearing resin-bound anticodons, which could sequence-specifically separate a population of DNA-templates into spatially distinct locations by hybridization. According to this split-and-pool protocol a peptide
Peptides are short chains of amino acids linked by peptide bonds. A polypeptide is a longer, continuous, unbranched peptide chain. Polypeptides that have a molecular mass of 10,000 Da or more are called proteins. Chains of fewer than twenty am ...
combinatorial library DNA-encoded of 106 members was generated.
DNA-templated synthesis
 In 2001 David Liu and co-workers showed that complementary DNA
In 2001 David Liu and co-workers showed that complementary DNA oligonucleotides
Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, research, and forensics. Commonly made in the laboratory by solid-phase chemical synthesis, these small fragments of nucleic aci ...
can be used to assist certain synthetic reactions, which do not efficiently take place in solution
Solution may refer to:
* Solution (chemistry), a mixture where one substance is dissolved in another
* Solution (equation), in mathematics
** Numerical solution, in numerical analysis, approximate solutions within specified error bounds
* Solu ...
at low concentration
In chemistry, concentration is the abundance of a constituent divided by the total volume of a mixture. Several types of mathematical description can be distinguished: '' mass concentration'', '' molar concentration'', '' number concentration'', ...
. A DNA-heteroduplex was used to accelerate the reaction between chemical moieties displayed at the extremities of the two DNA strands. Furthermore, the "proximity effect", which accelerates bimolecular reaction, was shown to be distance-independent (at least within a distance of 30 nucleotides
Nucleotides are Organic compound, organic molecules composed of a nitrogenous base, a pentose sugar and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both o ...
). In a sequence-programmed fashion oligonucleotides carrying one chemical reactant group were hybridized to complementary oligonucleotide derivatives carrying a different reactive chemical group. The proximity conferred by the DNA hybridization drastically increases the effective molarity
Molar concentration (also called molarity, amount concentration or substance concentration) is the number of moles of solute per liter of solution. Specifically, It is a measure of the concentration of a chemical species, in particular, of a solu ...
of the reaction reagents attached to the oligonucleotides, enabling the desired reaction to occur even in an aqueous environment at concentrations which are several orders of magnitude lower than those needed for the corresponding conventional organic reaction not DNA-templated. Using a DNA-templated set-up and sequence-programmed synthesis Liu and co-workers generated a 64-member compound DNA encoded library of macrocycles
Macrocycles are often described as molecules and ions containing a Ring (chemistry), ring of twelve or more atoms. Classical examples include the crown ethers, calixarenes, porphyrins, and cyclodextrins. Macrocycles describe a large, mature area ...
.
3-Dimensional proximity-based technology (YoctoReactor technology)
The YoctoReactor (yR) is a 3D proximity-driven approach which exploits the self-assembling nature of DNA oligonucleotides into 3, 4 or 5-way junctions to direct small molecule synthesis at the center of the junction. Figure 5 illustrates the basic concept with a 4-way DNA junction.yocto
A metric prefix is a unit prefix that precedes a basic unit of measure to indicate a multiple or submultiple of the unit. All metric prefixes used today are decadic. Each prefix has a unique symbol that is prepended to any unit symbol. The pre ...
liter, hence the name YoctoReactor. This volume contains a single molecule reaction yielding reaction concentrations in the high mM range. The effective concentration facilitated by the DNA greatly accelerates chemical reactions that otherwise would not take place at the actual concentration several orders of magnitude lower.
Building a yR library
Figure 6 illustrates the generation of a yR library using a 3-way DNA junction.Homogeneous screening of yoctoreactor libraries
A homogeneous method for screening yoctoreactor libraries (yR) has recently been developed which uses water-in-oil emulsion technology to isolate individual ligand-target complexes. Called Binder Trap Enrichment (BTE), ligands to a protein target are identified by trapping binding pairs (DNA-labelled protein target and yR ligand) in emulsion droplets during dissociation dominated kinetics. Once trapped, the target and ligand DNA are joined by ligation, thus preserving the binding information. Hereafter, identification of hits is essentially a counting exercise: information on binding events is deciphered by sequencing and counting the joined DNA - selective binders are counted with a much higher frequency than random binders. This is possible because random trapping of target and ligand is "diluted" by the high number of water droplets in the emulsion. The low noise and background signal characteristic of BTE is attributed to the "dilution" of the random signal, the lack of surface artifacts and the high fidelity of the yR library and screening method. Screening is performed in a single tube method. Biologically active hits are identified in a single round of BTE characterized by a low false positive rate. BTE mimics the non-equilibrium nature of in vivo ligand-target interactions and offers the unique possibility to screen for target specific ligands based on ligand-target residence time because the emulsion, which traps the binding complex, is formed during a dynamic dissociation phase.Screening of DELs in cells
DNA-encoded libraries (DELs) have been adapted for screening in living cells to better reflect native biological conditions, specifically using ''Xenopus laevis
The African clawed frog (''Xenopus laevis''), also known as simply xenopus, African clawed toad, African claw-toed frog or the ''platanna'') is a species of African aquatic frog of the family Pipidae. Its name is derived from the short black ...
'' oocytes. This approach, termed cellular Binder Trap Enrichment (cBTE), facilitates the identification of small-molecule ligands that bind to target proteins in a native cellular environment.
In this method, the protein of interest (POI) is expressed in oocytes as a fusion with a "Prey" protein, such as carbonic anhydrase IX (CAIX). Simultaneously, a "Bait" molecule—comprising a known ligand for the Prey protein linked to a DNA strand—is introduced. Alongside the Bait, a DEL is co-injected into the oocytes. If a DEL member binds to the POI, it brings its attached DNA tag into the same molecular complex as the Bait DNA via the POI–Prey–Bait interaction.
Following incubation, the oocytes are lysed, and the lysate is subjected to Binder Trap Enrichment (BTE) as described above. In essence, the DEL and Bait DNA are ligated in droplets, thus encoding the binding event. The ligated DNA is then amplified and subjected to high-throughput sequencing
DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, thymine, cytosine, and guanine. The ...
to identify the DEL members that interacted with the POI.
 This intracellular screening technique allows for the discovery of ligands that engage targets in their native conformation and cellular environment, enabling screening of targets that are difficult to express or purify, and potentially improving the physiological relevance of identified compounds.
This intracellular screening technique allows for the discovery of ligands that engage targets in their native conformation and cellular environment, enabling screening of targets that are difficult to express or purify, and potentially improving the physiological relevance of identified compounds.
Decoding of DNA-encoded chemical libraries
Following selection from DNA-encoded chemical libraries, the decoding strategy for the fast and efficient identification of the specific binding compounds is crucial for the further development of the DEL technology. So far, Sanger-sequencing-based decoding,microarray
A microarray is a multiplex (assay), multiplex lab-on-a-chip. Its purpose is to simultaneously detect the expression of thousands of biological interactions. It is a two-dimensional array on a Substrate (materials science), solid substrate—usu ...
-based methodology and high-throughput sequencing
DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, thymine, cytosine, and guanine. The ...
techniques represented the main methodologies for the decoding of DNA-encoded library selections.
Sanger sequencing-based decoding
Although many authors implicitly envisaged a traditionalSanger sequencing
Sanger sequencing is a method of DNA sequencing that involves electrophoresis and is based on the random incorporation of chain-terminating dideoxynucleotides by DNA polymerase during in vitro DNA replication. After first being developed by Fred ...
-based decoding, the number of codes to sequence simply according to the complexity of the library is definitely an unrealistic task for a traditional Sanger sequencing
Sanger sequencing is a method of DNA sequencing that involves electrophoresis and is based on the random incorporation of chain-terminating dideoxynucleotides by DNA polymerase during in vitro DNA replication. After first being developed by Fred ...
approach. Nevertheless, the implementation of Sanger sequencing
Sanger sequencing is a method of DNA sequencing that involves electrophoresis and is based on the random incorporation of chain-terminating dideoxynucleotides by DNA polymerase during in vitro DNA replication. After first being developed by Fred ...
for decoding DNA-encoded chemical libraries in high-throughput fashion was the first to be described. After selection and PCR amplification
The polymerase chain reaction (PCR) is a method widely used to make millions to billions of copies of a specific DNA sample rapidly, allowing scientists to amplify a very small sample of DNA (or a part of it) sufficiently to enable detailed st ...
of the DNA-tags of the library compounds, concatamers containing multiple coding sequences were generated and ligated into a vector
Vector most often refers to:
* Euclidean vector, a quantity with a magnitude and a direction
* Disease vector, an agent that carries and transmits an infectious pathogen into another living organism
Vector may also refer to:
Mathematics a ...
. Following Sanger sequencing
Sanger sequencing is a method of DNA sequencing that involves electrophoresis and is based on the random incorporation of chain-terminating dideoxynucleotides by DNA polymerase during in vitro DNA replication. After first being developed by Fred ...
of a representative number of the resulting colonies
A colony is a territory subject to a form of foreign rule, which rules the territory and its indigenous peoples separated from the foreign rulers, the colonizer, and their '' metropole'' (or "mother country"). This separated rule was often or ...
revealed the frequencies of the codes present in the DNA-encoded library sample before and after selection.
Microarray-based decoding
A DNAmicroarray
A microarray is a multiplex (assay), multiplex lab-on-a-chip. Its purpose is to simultaneously detect the expression of thousands of biological interactions. It is a two-dimensional array on a Substrate (materials science), solid substrate—usu ...
is a device for high-throughput investigations widely used in molecular biology
Molecular biology is a branch of biology that seeks to understand the molecule, molecular basis of biological activity in and between Cell (biology), cells, including biomolecule, biomolecular synthesis, modification, mechanisms, and interactio ...
and in medicine
Medicine is the science and Praxis (process), practice of caring for patients, managing the Medical diagnosis, diagnosis, prognosis, Preventive medicine, prevention, therapy, treatment, Palliative care, palliation of their injury or disease, ...
. It consists of an arrayed series of microscopic spots (‘features’ or ‘locations’) containing few picomoles of oligonucleotides
Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, research, and forensics. Commonly made in the laboratory by solid-phase chemical synthesis, these small fragments of nucleic aci ...
carrying a specific DNA sequence. This can be a short section of a gene
In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protei ...
or other DNA element that are used as probes to hybridize a DNA or RNA
Ribonucleic acid (RNA) is a polymeric molecule that is essential for most biological functions, either by performing the function itself (non-coding RNA) or by forming a template for the production of proteins (messenger RNA). RNA and deoxyrib ...
sample under suitable conditions. Probe-target hybridization is usually detected and quantified by fluorescence
Fluorescence is one of two kinds of photoluminescence, the emission of light by a substance that has absorbed light or other electromagnetic radiation. When exposed to ultraviolet radiation, many substances will glow (fluoresce) with colore ...
-based detection of fluorophore
A fluorophore (or fluorochrome, similarly to a chromophore) is a fluorescent chemical compound that can re-emit light upon light excitation. Fluorophores typically contain several combined aromatic groups, or planar or cyclic molecules with se ...
-labeled targets to determine relative abundance of the target nucleic acid
Nucleic acids are large biomolecules that are crucial in all cells and viruses. They are composed of nucleotides, which are the monomer components: a pentose, 5-carbon sugar, a phosphate group and a nitrogenous base. The two main classes of nuclei ...
sequences. Microarray
A microarray is a multiplex (assay), multiplex lab-on-a-chip. Its purpose is to simultaneously detect the expression of thousands of biological interactions. It is a two-dimensional array on a Substrate (materials science), solid substrate—usu ...
has been used for the successfully decoding of ESAC DNA-encoded libraries and PNA-encoded libraries. The coding oligonucleotides
Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, research, and forensics. Commonly made in the laboratory by solid-phase chemical synthesis, these small fragments of nucleic aci ...
representing the individual chemical compounds in the library, are spotted and chemically linked onto the microarray
A microarray is a multiplex (assay), multiplex lab-on-a-chip. Its purpose is to simultaneously detect the expression of thousands of biological interactions. It is a two-dimensional array on a Substrate (materials science), solid substrate—usu ...
slides, using a BioChip Arrayer robot. Subsequently, the oligonucleotide
Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, Recombinant DNA, research, and Forensic DNA, forensics. Commonly made in the laboratory by Oligonucleotide synthesis, solid-phase ...
tags of the binding compounds isolated from the selection are PCR amplified using a fluorescent
Fluorescence is one of two kinds of photoluminescence, the emission of light by a substance that has absorbed light or other electromagnetic radiation. When exposed to ultraviolet radiation, many substances will glow (fluoresce) with color ...
primer and hybridized onto the DNA-microarray
A microarray is a multiplex (assay), multiplex lab-on-a-chip. Its purpose is to simultaneously detect the expression of thousands of biological interactions. It is a two-dimensional array on a Substrate (materials science), solid substrate—usu ...
slide. Afterwards, microarrays
A microarray is a multiplex lab-on-a-chip. Its purpose is to simultaneously detect the expression of thousands of biological interactions. It is a two-dimensional array on a solid substrate—usually a glass slide or silicon thin-film cell� ...
are analyzed using a laser
A laser is a device that emits light through a process of optical amplification based on the stimulated emission of electromagnetic radiation. The word ''laser'' originated as an acronym for light amplification by stimulated emission of radi ...
scan and spot intensities detected and quantified. The enrichment of the preferential binding compounds is revealed comparing the spots intensity of the DNA-microarray
A microarray is a multiplex (assay), multiplex lab-on-a-chip. Its purpose is to simultaneously detect the expression of thousands of biological interactions. It is a two-dimensional array on a Substrate (materials science), solid substrate—usu ...
slide before and after selection.
Decoding by high throughput sequencing
According to the complexity of the DNA encoded chemical library (typically between 103 and 106 members), a conventionalSanger sequencing
Sanger sequencing is a method of DNA sequencing that involves electrophoresis and is based on the random incorporation of chain-terminating dideoxynucleotides by DNA polymerase during in vitro DNA replication. After first being developed by Fred ...
based decoding is unlikely to be usable in practice, due both to the high cost per base for the sequencing and to the tedious procedure involved. High throughput sequencing
DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, thymine, cytosine, and guanine. The ...
technologies exploited strategies that parallelize the sequencing process displacing the use of capillary
A capillary is a small blood vessel, from 5 to 10 micrometres in diameter, and is part of the microcirculation system. Capillaries are microvessels and the smallest blood vessels in the body. They are composed of only the tunica intima (the inn ...
electrophoresis
Electrophoresis is the motion of charged dispersed particles or dissolved charged molecules relative to a fluid under the influence of a spatially uniform electric field. As a rule, these are zwitterions with a positive or negative net ch ...
and producing thousands or millions of sequences at once. In 2008 was described the first implementation of a high-throughput sequencing
DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, thymine, cytosine, and guanine. The ...
technique originally developed for genome sequencing (i.e. " 454 technology") to the fast and efficient decoding of a DNA encoded chemical library comprising 4000 compounds. This study led to the identification of novel chemical compounds with submicromolar dissociation constant
In chemistry, biochemistry, and pharmacology, a dissociation constant (''K''D) is a specific type of equilibrium constant that measures the propensity of a larger object to separate (dissociate) reversibly into smaller components, as when a complex ...
s towards streptavidin
Streptavidin is a 52 Atomic mass unit, kDa protein (tetramer) purified from the bacterium ''Streptomyces avidinii''. Streptavidin Homotetramer, homo-tetramers have an extraordinarily high affinity for biotin (also known as vitamin B7 or vitamin ...
and definitely shown the feasibility to construct, perform selections and decode DNA-encoded libraries containing millions of chemical compounds.
See also
*Drug discovery
In the fields of medicine, biotechnology, and pharmacology, drug discovery is the process by which new candidate medications are discovered.
Historically, drugs were discovered by identifying the active ingredient from traditional remedies or ...
* High-throughput screening
High-throughput screening (HTS) is a method for scientific discovery especially used in drug discovery and relevant to the fields of biology, materials science and chemistry. Using robotics, data processing/control software, liquid handling device ...
* Combinatorial chemistry
Combinatorial chemistry comprises chemical synthesis, chemical synthetic methods that make it possible to prepare a large number (tens to thousands or even millions) of compounds in a single process. These compound library, compound libraries can b ...
* DNA sequencing
DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, thymine, cytosine, and guanine. The ...
* Phage display
Phage display is a laboratory technique for the study of protein–protein, protein–peptide, and protein–DNA interactions that uses bacteriophages (viruses that infect bacteria) to connect proteins with the genetic information that encodes ...
References
{{DEFAULTSORT:Dna Encoded Chemical Library Biotechnology Scientific techniques Drug discovery Molecular biology Combinatorial chemistry