|
HMGN
HMGN (High Mobility Group Nucleosome-binding) proteins are members of the broader class of high mobility group (HMG) chromosomal proteins that are involved in regulation of Transcription (biology), transcription, DNA replication, replication, Genetic recombination, recombination, and DNA repair. HMGN1 and HMGN2 (initially designated HMG-14 and HMG-17 respectively) were discovered by E.W. Johns research group in the early 1970s. HMGN3, HMGN4, and HMGN5 were discovered later and are less abundant. HMGNs are nucleosome binding proteins that help in transcription, replication, recombination, and DNA repair. They can also alter the chromatin epigenetic landscape, helping to stabilize cell identity. There is still relatively little known about their structure and function. HMGN proteins are found in all vertebrates, and play a role in chromatin structure and histone modification. HMGNs come in long chains of amino acids, containing around 100 for HMGN1-4, and roughly 200 in HMGN5. Rec ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hmgn
HMGN (High Mobility Group Nucleosome-binding) proteins are members of the broader class of high mobility group (HMG) chromosomal proteins that are involved in regulation of Transcription (biology), transcription, DNA replication, replication, Genetic recombination, recombination, and DNA repair. HMGN1 and HMGN2 (initially designated HMG-14 and HMG-17 respectively) were discovered by E.W. Johns research group in the early 1970s. HMGN3, HMGN4, and HMGN5 were discovered later and are less abundant. HMGNs are nucleosome binding proteins that help in transcription, replication, recombination, and DNA repair. They can also alter the chromatin epigenetic landscape, helping to stabilize cell identity. There is still relatively little known about their structure and function. HMGN proteins are found in all vertebrates, and play a role in chromatin structure and histone modification. HMGNs come in long chains of amino acids, containing around 100 for HMGN1-4, and roughly 200 in HMGN5. Rec ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
High Mobility Group
High-Mobility Group or HMG is a group of chromosomal proteins that are involved in the regulation of DNA-dependent processes such as transcription, replication, recombination, and DNA repair. Families The HMG proteins are subdivided into 3 superfamilies each containing a characteristic functional domain: * HMGA – contains an AT-hook domain ** HMGA1 ** HMGA2 * HMGB – contains a HMG-box domain ** HMGB1 ** HMGB2 ** HMGB3 ** HMGB4 * HMGN – contains a nucleosomal binding domain ** HMGN1 ** HMGN2 ** HMGN3 ** HMGN4 ** HMGN5 Proteins containing any of these embedded in their sequence are known as HMG motif proteins. HMG-box proteins are found in a variety of eukaryotic organisms. They were originally isolated from mammalian cells, and named according to their electrophoretic mobility in polyacrylamide gels. Other families with HMG-box domain * SOX gene family ** Sex-Determining Region Y Protein ** SOX1, SOX2, etc. * TCF/LEF family (T cell factor/lymphoid enhancer f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mouse Embryonic Fibroblast
Mouse Embryonic Fibroblasts (MEFs) are a type of fibroblast prepared from mouse embryo. MEFs show a spindle shape when cultured ''in vitro'', a typical feature of fibroblasts. The MEF is a limited cell line. After several transmission, MEFs will senesce and finally die off. Nevertheless, researchers can use several strategies, like virus infection or repeated transmission to immortalize MEF cells, which can let MEFs grown indefinitely in spite of some changes in characters. MEFs are widely used in life science researches, especially in stem cell biology. Preparation and culture To prepare MEFs, pregnant female mice are needed. After killing the female mouse, the researcher should incise its stomach and then detach the embryo from the placenta in a biohazard hood. Then the liver and head should be taken out. Finally digest the remains by enzymes to obtain single isolated cells and culture the cells in a tissue culture dishes. MEF cells can be cultured ''in vitro'' in DMEM mediu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
H3K4me1
H3K4me1 is an epigenetic modification to the DNA packaging protein Histone H3. It is a mark that indicates the mono-methylation at the 4th lysine residue of the histone H3 protein and often associated with gene enhancers. Nomenclature H3K4me1 indicates monomethylation of lysine 4 on histone H3 protein subunit: Lysine methylation This diagram shows the progressive methylation of a lysine residue. The mono-methylation denotes the methylation present in H3K4me1. Understanding histone modifications The genomic DNA of eukaryotic cells is wrapped around special protein molecules known as histones. The complexes formed by the looping of the DNA are known as chromatin. The basic structural unit of chromatin is the nucleosome: this consists of the core octamer of histones (H2A, H2B, H3 and H4) as well as a linker histone and about 180 base pairs of DNA. These core histones are rich in lysine and arginine residues. The carboxyl (C) terminal end of these histones contribute to his ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
H3K27ac
H3K27ac is an epigenetic modification to the DNA packaging protein histone H3. It is a mark that indicates acetylation of the lysine residue at N-terminal position 27 of the histone H3 protein. H3K27ac is associated with the higher activation of transcription and therefore defined as an ''active enhancer'' mark. H3K27ac is found at both proximal and distal regions of transcription start site (TSS). Lysine acetylation and deacetylation Proteins are typically acetylated on lysine residues, and the acetylation reaction relies on acetyl-coenzyme A as the acetyl group donor. In histone acetylation and deacetylation, histone proteins are acetylated and deacetylated on lysine residues in the N-terminal tail as part of gene regulation. Typically, these reactions are catalyzed by enzymes with ''histone acetyltransferase'' (HAT) or ''histone deacetylase'' (HDAC) activity, although HATs and HDACs can modify the acetylation status of non-histone proteins as well. The regulation of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
H3K27me3
H3K27me3 is an epigenetic modification to the DNA packaging protein Histone H3. It is a mark that indicates the tri-methylation of lysine 27 on histone H3 protein. This tri-methylation is associated with the downregulation of nearby genes via the formation of heterochromatic regions. Nomenclature H3K27me3 indicates trimethylation of lysine 27 on histone H3 protein subunit: Lysine methylation This diagram shows the progressive methylation of a lysine residue. The tri-methylation denotes the methylation present in H3K27me3. Understanding histone modifications The genomic DNA of eukaryotic cells is wrapped around special protein molecules known as Histones. The complexes formed by the looping of the DNA are known as Chromatin. The basic structural unit of chromatin is the Nucleosome: this consists of the core octamer of histones (H2A, H2B, H3 and H4) as well as a linker histone and about 180 base pairs of DNA. These core histones are rich in lysine and arginine residues. The ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ChIP-sequencing
ChIP-sequencing, also known as ChIP-seq, is a method used to analyze protein interactions with DNA. ChIP-seq combines chromatin immunoprecipitation (ChIP) with massively parallel DNA sequencing to identify the binding sites of DNA-associated proteins. It can be used to map global binding sites precisely for any protein of interest. Previously, ChIP-on-chip was the most common technique utilized to study these protein–DNA relations. Uses ChIP-seq is primarily used to determine how transcription factors and other chromatin-associated proteins influence phenotype-affecting mechanisms. Determining how proteins interact with DNA to regulate gene expression is essential for fully understanding many biological processes and disease states. This epigenetic information is complementary to genotype and expression analysis. ChIP-seq technology is currently seen primarily as an alternative to ChIP-chip which requires a hybridization array. This introduces some bias, as an array is restri ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Kinase C
In cell biology, Protein kinase C, commonly abbreviated to PKC (EC 2.7.11.13), is a family of protein kinase enzymes that are involved in controlling the function of other proteins through the phosphorylation of hydroxyl groups of serine and threonine amino acid residues on these proteins, or a member of this family. PKC enzymes in turn are activated by signals such as increases in the concentration of diacylglycerol (DAG) or calcium ions (Ca2+). Hence PKC enzymes play important roles in several signal transduction cascades. In biochemistry, the PKC family consists of fifteen isozymes in humans. They are divided into three subfamilies, based on their second messenger requirements: conventional (or classical), novel, and atypical. Conventional (c)PKCs contain the isoforms α, βI, βII, and γ. These require Ca2+, DAG, and a phospholipid such as phosphatidylserine for activation. Novel (n)PKCs include the δ, ε, η, and θ isoforms, and require DAG, but do not require Ca2+ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
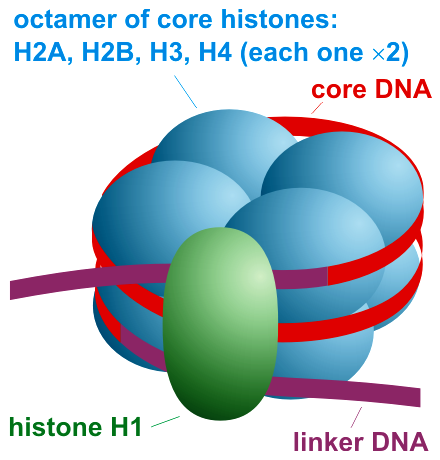
Histone H1
Histone H1 is one of the five main histone protein families which are components of chromatin in eukaryotic cells. Though highly conserved, it is nevertheless the most variable histone in sequence across species. Structure Metazoan H1 proteins feature a central globular "winged helix" domain and long C- and short N-terminal tails. H1 is involved with the packing of the "beads on a string" sub-structures into a high order structure, whose details have not yet been solved. H1 found in protists and bacteria, otherwise known as nucleoproteins HC1 and HC2 (, ), lack the central domain and the N-terminal tail. H1 is less conserved than core histones. The globular domain is the most conserved part of H1. Function Unlike the other histones, H1 does not make up the nucleosome "bead". Instead, it sits on top of the structure, keeping in place the DNA that has wrapped around the nucleosome. H1 is present in half the amount of the other four histones, which contribute two molecules ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleosome Organization
A nucleosome is the basic structural unit of DNA packaging in eukaryotes. The structure of a nucleosome consists of a segment of DNA wound around eight histone proteins and resembles thread wrapped around a spool. The nucleosome is the fundamental subunit of chromatin. Each nucleosome is composed of a little less than two turns of DNA wrapped around a set of eight proteins called histones, which are known as a histone octamer. Each histone octamer is composed of two copies each of the histone proteins H2A, H2B, H3, and H4. DNA must be compacted into nucleosomes to fit within the cell nucleus. In addition to nucleosome wrapping, eukaryotic chromatin is further compacted by being folded into a series of more complex structures, eventually forming a chromosome. Each human cell contains about 30 million nucleosomes. Nucleosomes are thought to carry epigenetically inherited information in the form of covalent modifications of their core histones. Nucleosome positions in the geno ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |