translocase on:
[Wikipedia]
[Google]
[Amazon]
Translocase is a general term for a
 This ATPase carries out the
This ATPase carries out the
 ;Protein import into mitochondria: Hundreds of proteins encoded by the nucleus are required for mitochondrial metabolism, growth, division, and partitioning to daughter cells, and all of these proteins must be imported into the organelle. Translocase of the outer membrane (TOM) and translocase of the inner membrane (TIM) mediate the import of proteins into the mitochondrion. The translocase of the outer membrane (TOM) sorts proteins via several mechanisms either directly to the outer membrane, the intermembrane space, or the translocase of the inner membrane (TIM). Then, generally, the TIM23 machinery mediates protein translocation into the matrix and the TIM22 machinery mediates insertion into the inner membrane.
;Fatty acids import into mitochondria (Carnitine Shuttle System): Carnitine-acylcarnitine translocase (CACT) catalyzes both unidirectional transport of carnitine and carnitine/acylcarnitine exchange in the inner mitochondrial membrane, allowing the import of long-chain fatty acids into the mitochondria where they are oxidized by the
;Protein import into mitochondria: Hundreds of proteins encoded by the nucleus are required for mitochondrial metabolism, growth, division, and partitioning to daughter cells, and all of these proteins must be imported into the organelle. Translocase of the outer membrane (TOM) and translocase of the inner membrane (TIM) mediate the import of proteins into the mitochondrion. The translocase of the outer membrane (TOM) sorts proteins via several mechanisms either directly to the outer membrane, the intermembrane space, or the translocase of the inner membrane (TIM). Then, generally, the TIM23 machinery mediates protein translocation into the matrix and the TIM22 machinery mediates insertion into the inner membrane.
;Fatty acids import into mitochondria (Carnitine Shuttle System): Carnitine-acylcarnitine translocase (CACT) catalyzes both unidirectional transport of carnitine and carnitine/acylcarnitine exchange in the inner mitochondrial membrane, allowing the import of long-chain fatty acids into the mitochondria where they are oxidized by the
 This subclass contains translocases that catalyze the translocation of hydrons. Based on the reaction they are linked to, EC 7.1 can be further classified into:
This subclass contains translocases that catalyze the translocation of hydrons. Based on the reaction they are linked to, EC 7.1 can be further classified into:
EC 7.1.1
Hydron translocation or charge separation linked to oxidoreductase reactions
EC 7.1.2
Hydron translocation linked to the
EC 7.1.3
Hydron translocation linked to the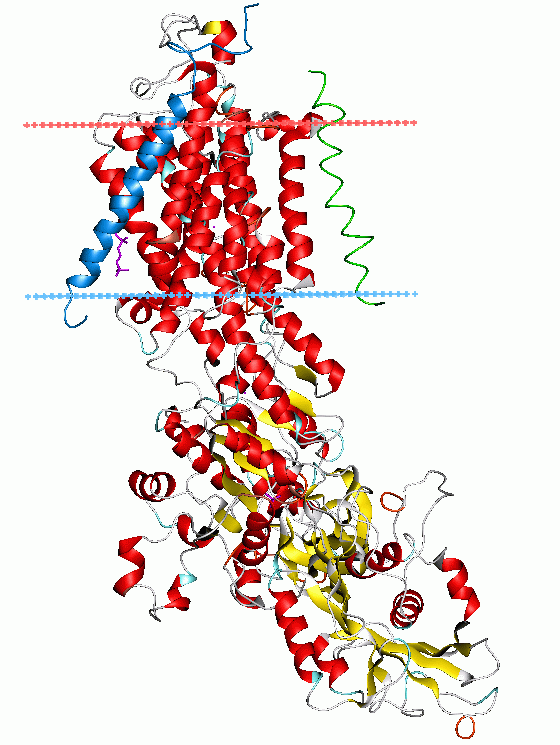
EC 7.2.1
Translocation of inorganic cations linked to oxidoreductase reactions
EC 7.2.2
Translocation of inorganic cations linked to the hydrolysis of a nucleoside triphosphate
EC 7.2.4
Translocation of inorganic cations linked to decarboxylation An important translocase contained in this group is Na+/K+ pump, also known as EC 7.2.2.13.
EC 7.3.2
Translocation of inorganic anions linked to the hydrolysis of a nucleoside triphosphate.
7.3.2.1
ABC-type phosphate transporter: The expected taxonomic range for this enzyme is: Eukaryota, Bacteria. A bacterial enzyme that interacts with an extracytoplasmic substrate binding protein and mediates the high affinity uptake of phosphate anions. Unlike P-type ATPases, it does not undergo
7.3.2.2
ABC-type phosphonate transporter: The enzyme, found in bacteria, interacts with an extracytoplasmic substrate binding protein and mediates the import of phosphonate and organophosphate anions. :* ATP + H2O + phosphonate hosphonate-binding proteinside 1] = ADP + phosphate + phosphonate ide 2+ hosphonate- binding proteinside 1]
7.3.2.3
ABC-type sulfate transporter: The expected taxonomic range for this enzyme is: Eukaryota, Bacteria. The enzyme from
7.3.2.4
ABC-type nitrate transporter: The expected taxonomic range for this enzyme is: Eukaryota, Bacteria. The enzyme, found in bacteria, interacts with an extracytoplasmic substrate binding protein and mediates the import of nitrate, nitrite, and cyanate. :* ATP + H2O + nitrate itrate - binding proteinide 1= ADP + phosphate + nitrate ide 2+ itrate - binding proteinside 1]
7.3.2.5
ABC-type molybdate transporter: The expected taxonomic range for this enzyme is: Archaea, Eukaryota, Bacteria. The enzyme, found in bacteria, interacts with an extracytoplasmic substrate binding protein and mediates the high-affinity import of molybdate and
7.3.2.6
ABC-type tungstate transporter: The expected taxonomic range for this enzyme is: Archaea, Bacteria. The enzyme, characterized from the archaeon
7.4.2
Translocation of amino acids and peptides linked to the hydrolysis of a nucleoside triphosphate.
7.4.2.1
ABC-type polar-amino-acid transporter: The expected taxonomic range for this enzyme is: Eukaryota, Bacteria. The enzyme, found in bacteria, interacts with an extracytoplasmic substrate binding protein and mediates the import of polar amino acids. This entry comprises bacterial enzymes that import
7.4.2.2
ABC-type nonpolar-amino-acid transporter: The expected taxonomic range for this enzyme is: Eukaryota, Bacteria. The enzyme, found in bacteria, interacts with an extracytoplasmic substrate binding protein. This entry comprises enzymes that import
7.4.2.3
ABC-type mitochondrial protein-transporting ATPase: The expected taxonomic range for this enzyme is: Eukaryota, Bacteria. A non-phosphorylated, non-ABC (ATP-binding cassette) ATPase involved in the transport of proteins or preproteins into mitochondria using the TIM ( Translocase of the inner membrane, Translocase of the Inner Membrane) protein complex. TIM is the protein transport machinery of the mitochondrial inner membrane that contains three essential TIM proteins: Tim17 and Tim23 are thought to build a preprotein translocation channel while Tim44 interacts transiently with the matrix heat-shock protein
7.4.2.4
ABC-type chloroplast protein-transporting ATPase: The enzyme appears in viruses and cellular organisms. Involved in the transport of proteins or preproteins into chloroplast stroma (several ATPases may participate in this process). :* ATP + H2O + chloroplast protein ide 1= ADP + phosphate + chloroplast protein ide 2
7.4.2.5
ABC-type protein transporter: The expected taxonomic range for this enzyme is: Eukaryota, Bacteria. This entry stands for a family of bacterial enzymes that are dedicated to the secretion of one or several closely related proteins belonging to the toxin,
7.4.2.6
ABC-type oligopeptide transporter: A bacterial enzyme that interacts with an extracytoplasmic substrate binding protein and mediates the import of
7.4.2.7
ABC-type alpha-factor-pheromone transporter: The enzyme appears in viruses and cellular organisms characterized by the presence of two similar ATP-binding domains/proteins and two integral membrane domains/proteins. Does not undergo phosphorylation during the transport process. A yeast enzyme that exports the α-factor sex pheromone. :* ATP + H2O + alpha factor ide 1= ADP + phosphate + alpha factor ide 2
7.4.2.8
ABC-type protein-secreting ATPase: The expected taxonomic range for this enzyme is: Archaea, Bacteria. A non-phosphorylated, non-ABC (ATP-binding cassette) ATPase that is involved in protein transport. :* ATP + H2O + cellular protein ide 1= ADP + phosphate + cellular protein ide 2
7.4.2.9
ABC-type dipeptide transporter: The enzyme appears in viruses and cellular organisms. ATP-binding cassette (ABC) type transporter, characterized by the presence of two similar ATP-binding domains/proteins and two integral membrane domains/proteins. A bacterial enzyme that interacts with an extracytoplasmic substrate binding protein and mediates the uptake of dipeptides and tripeptides. :* ATP + H2O + dipeptide ipeptide - binding proteinide 1= ADP + phosphate + ide 2+ ipeptide - binding proteinside 1]
7.4.2.10
ABC-type glutathione transporter: A prokaryotic ATP-binding cassette (ABC) type transporter, characterized by the presence of two similar ATP-binding domains/proteins and two integral membrane domains/proteins. The enzyme from the bacterium ''
7.4.2.11
ABC-type methionine transporter: A bacterial enzyme that interacts with an extracytoplasmic substrate binding protein and functions to import
7.4.2.12
ABC-type cystine transporter: A bacterial enzyme that interacts with an extracytoplasmic substrate binding protein and mediates the high affinity import of trace cystine. The enzyme from ''
EC 7.5.2
Linked to the hydrolysis of a
EC 7.6.2
Linked to the hydrolysis of a
protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, res ...
that assists in moving another molecule
A molecule is a group of two or more atoms held together by attractive forces known as chemical bonds; depending on context, the term may or may not include ions which satisfy this criterion. In quantum physics, organic chemistry, and b ...
, usually across a cell membrane. These enzymes
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. ...
catalyze the movement of ions or molecules across membranes or their separation within membranes. The reaction is designated as a transfer from “side 1” to “side 2” because the designations “in” and “out”, which had previously been used, can be ambiguous. Translocases are the most common secretion system file:Secretory mechanism.jpg, 440px
Secretion is the movement of material from one point to another, such as a secreted chemical substance from a cell (biology), cell or gland. In contrast, excretion is the removal of certain substances or waste pr ...
in Gram positive
In bacteriology, gram-positive bacteria are bacteria that give a positive result in the Gram stain test, which is traditionally used to quickly classify bacteria into two broad categories according to their type of cell wall.
Gram-positive bact ...
bacteria
Bacteria (; singular: bacterium) are ubiquitous, mostly free-living organisms often consisting of one biological cell. They constitute a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria were am ...
.
It is also a historical term for the protein now called elongation factor G, due to its function in moving the transfer RNA
Transfer RNA (abbreviated tRNA and formerly referred to as sRNA, for soluble RNA) is an adaptor molecule composed of RNA, typically 76 to 90 nucleotides in length (in eukaryotes), that serves as the physical link between the mRNA and the amino ...
(tRNA) and messenger RNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
mRNA is created during the ...
(mRNA) through the ribosome
Ribosomes ( ) are macromolecular machines, found within all cells, that perform biological protein synthesis (mRNA translation). Ribosomes link amino acids together in the order specified by the codons of messenger RNA (mRNA) molecules to fo ...
.
History
The enzyme classification and nomenclature list was first approved by theInternational Union of Biochemistry
The International Union of Biochemistry and Molecular Biology (IUBMB) is an international non-governmental organisation concerned with biochemistry and molecular biology. Formed in 1955 as the International Union of Biochemistry (IUB), the union ...
in 1961. Six enzyme classes had been recognized based on the type of chemical reaction catalyzed, including oxidoreductases (EC 1), transferases (EC 2), hydrolases (EC 3), lyases
In biochemistry, a lyase is an enzyme that catalyzes the breaking (an elimination reaction) of various chemical bonds by means other than hydrolysis (a substitution reaction) and oxidation, often forming a new double bond or a new ring structu ...
(EC 4), isomerases (EC 5) and ligases
In biochemistry, a ligase is an enzyme that can catalyze the joining ( ligation) of two large molecules by forming a new chemical bond. This is typically via hydrolysis of a small pendant chemical group on one of the larger molecules or the enzy ...
(EC 6). However, it became apparent that none of these could describe the important group of enzymes that catalyse the movement of ions or molecules across membranes or their separation within membranes. Several of these involve the hydrolysis of ATP and had been previously classified as ATPases (EC 3.6.3.-), although the hydrolytic reaction is not their primary function. In August 2018, the International Union of Biochemistry and Molecular Biology classified these enzymes under a new enzyme class (EC) of translocases (EC 7).
Mechanism of catalysis
The reaction most translocases catalyse is: * AX + Bside 1, , = A + X + , , Bside 2 A clear example of an enzyme that follows this scheme is H+-transporting two-sector ATPase: * ATP + H2O + 4 H+side 1 = ADP + phosphate + 4 H+side 2 This ATPase carries out the
This ATPase carries out the dephosphorylation
In biochemistry, dephosphorylation is the removal of a phosphate (PO43−) group from an organic compound by hydrolysis. It is a reversible post-translational modification. Dephosphorylation and its counterpart, phosphorylation, activate and de ...
of ATP into ADP while it transports H+ to the other side of the membrane.
However, other enzymes that also fall into this category do not follow the same reaction scheme. This is the case of ascorbate ferrireductase:
* ascorbateside 1 + Fe(III)side 2 = monodehydroascorbateside 1 + Fe(II)side 2
In which the enzyme only transports an electron in the catalysation of an oxidoreductase reaction between a molecule and an inorganic cation located on different sides of the membrane.
Function
The basic function, as already mentioned (see: ), is to "catalyse the movement of ions or molecules across membranes or their separation within membranes". This form ofmembrane transport
In cellular biology, membrane transport refers to the collection of mechanisms that regulate the passage of solutes such as ions and small molecules through biological membranes, which are lipid bilayers that contain proteins embedded in them. The ...
is classified under active membrane transport, an energy-requiring process of pumping molecules and ions across membranes against a concentration gradient.
Translocases biological importance relies primarily on their critical function, in the way that they provide movement across the cell's membrane in many cellular processes that are substantial, such as:
;Oxidative phosphorylation: ADP/ATP translocase (ANT) imports adenosine diphosphate ADP from the cytosol and exports ATP from the mitochondrial matrix, which are key transport steps for oxidative phosphorylation
Oxidative phosphorylation (UK , US ) or electron transport-linked phosphorylation or terminal oxidation is the metabolic pathway in which cells use enzymes to oxidize nutrients, thereby releasing chemical energy in order to produce adenosine t ...
in eukaryotic organisms. ADP from the cytosol is transported back into the mitochondrion for ATP synthesis and the synthesised ATP, produced from oxidative phosphorylation, is exported out of the mitochondrion for use in the cytosol, providing the cells with its main energy currency.
 ;Protein import into mitochondria: Hundreds of proteins encoded by the nucleus are required for mitochondrial metabolism, growth, division, and partitioning to daughter cells, and all of these proteins must be imported into the organelle. Translocase of the outer membrane (TOM) and translocase of the inner membrane (TIM) mediate the import of proteins into the mitochondrion. The translocase of the outer membrane (TOM) sorts proteins via several mechanisms either directly to the outer membrane, the intermembrane space, or the translocase of the inner membrane (TIM). Then, generally, the TIM23 machinery mediates protein translocation into the matrix and the TIM22 machinery mediates insertion into the inner membrane.
;Fatty acids import into mitochondria (Carnitine Shuttle System): Carnitine-acylcarnitine translocase (CACT) catalyzes both unidirectional transport of carnitine and carnitine/acylcarnitine exchange in the inner mitochondrial membrane, allowing the import of long-chain fatty acids into the mitochondria where they are oxidized by the
;Protein import into mitochondria: Hundreds of proteins encoded by the nucleus are required for mitochondrial metabolism, growth, division, and partitioning to daughter cells, and all of these proteins must be imported into the organelle. Translocase of the outer membrane (TOM) and translocase of the inner membrane (TIM) mediate the import of proteins into the mitochondrion. The translocase of the outer membrane (TOM) sorts proteins via several mechanisms either directly to the outer membrane, the intermembrane space, or the translocase of the inner membrane (TIM). Then, generally, the TIM23 machinery mediates protein translocation into the matrix and the TIM22 machinery mediates insertion into the inner membrane.
;Fatty acids import into mitochondria (Carnitine Shuttle System): Carnitine-acylcarnitine translocase (CACT) catalyzes both unidirectional transport of carnitine and carnitine/acylcarnitine exchange in the inner mitochondrial membrane, allowing the import of long-chain fatty acids into the mitochondria where they are oxidized by the β-oxidation
In biochemistry and metabolism, beta-oxidation is the catabolic process by which fatty acid molecules are broken down in the cytosol in prokaryotes and in the mitochondria in eukaryotes to generate acetyl-CoA, which enters the citric acid cyc ...
pathway. The mitochondrial membrane is impermeable to long-chain fatty acids, hence the need for this translocation.
Classification
The enzyme subclasses designate the types of components that are being transferred, and the sub-subclasses indicate the reaction processes that provide the driving force for the translocation.EC 7.1 Catalysing the translocation of hydrons
EC 7.1.1
Hydron translocation or charge separation linked to oxidoreductase reactions
EC 7.1.2
Hydron translocation linked to the
hydrolysis
Hydrolysis (; ) is any chemical reaction in which a molecule of water breaks one or more chemical bonds. The term is used broadly for substitution, elimination, and solvation reactions in which water is the nucleophile.
Biological hydrolysi ...
of a nucleoside triphosphate
A nucleoside triphosphate is a nucleoside containing a nitrogenous base bound to a 5-carbon sugar (either ribose or deoxyribose), with three phosphate groups bound to the sugar. They are the molecular precursors of both DNA and RNA, which ar ...
EC 7.1.3
Hydron translocation linked to the
hydrolysis
Hydrolysis (; ) is any chemical reaction in which a molecule of water breaks one or more chemical bonds. The term is used broadly for substitution, elimination, and solvation reactions in which water is the nucleophile.
Biological hydrolysi ...
of diphosphate
An important translocase contained in this group is ATP synthase
ATP synthase is a protein that catalyzes the formation of the energy storage molecule adenosine triphosphate (ATP) using adenosine diphosphate (ADP) and inorganic phosphate (Pi). It is classified under ligases as it changes ADP by the formation ...
, also known as EC 7.1.2.2.

EC 7.2 Catalysing the translocation of inorganic cations and their chelates
This subclass contains translocases that transferinorganic
In chemistry, an inorganic compound is typically a chemical compound that lacks carbon–hydrogen bonds, that is, a compound that is not an organic compound. The study of inorganic compounds is a subfield of chemistry known as ''inorganic chemist ...
cations
An ion () is an atom or molecule with a net electrical charge.
The charge of an electron is considered to be negative by convention and this charge is equal and opposite to the charge of a proton, which is considered to be positive by con ...
(metal cations). Based on the reaction they're linked to, EC 7.2 can be further classified into:
EC 7.2.1
Translocation of inorganic cations linked to oxidoreductase reactions
EC 7.2.2
Translocation of inorganic cations linked to the hydrolysis of a nucleoside triphosphate
EC 7.2.4
Translocation of inorganic cations linked to decarboxylation An important translocase contained in this group is Na+/K+ pump, also known as EC 7.2.2.13.
EC 7.3 Catalysing the translocation of inorganic anions
This subclass contains translocases that transfer inorganic cations anions. Subclasses are based on the reaction processes that provide the driving force for the translocation. At present only one subclass is representedEC 7.3.2
Translocation of inorganic anions linked to the hydrolysis of a nucleoside triphosphate.
7.3.2.1
ABC-type phosphate transporter: The expected taxonomic range for this enzyme is: Eukaryota, Bacteria. A bacterial enzyme that interacts with an extracytoplasmic substrate binding protein and mediates the high affinity uptake of phosphate anions. Unlike P-type ATPases, it does not undergo
phosphorylation
In chemistry, phosphorylation is the attachment of a phosphate group to a molecule or an ion. This process and its inverse, dephosphorylation, are common in biology and could be driven by natural selection. Text was copied from this source, wh ...
during the transport process.
:* ATP + H2O + phosphate hosphate - binding proteinside 1] = ADP + phosphate + phosphate ide 2+ hophate - binding proteinside 1]
7.3.2.2
ABC-type phosphonate transporter: The enzyme, found in bacteria, interacts with an extracytoplasmic substrate binding protein and mediates the import of phosphonate and organophosphate anions. :* ATP + H2O + phosphonate hosphonate-binding proteinside 1] = ADP + phosphate + phosphonate ide 2+ hosphonate- binding proteinside 1]
7.3.2.3
ABC-type sulfate transporter: The expected taxonomic range for this enzyme is: Eukaryota, Bacteria. The enzyme from
Escherichia coli
''Escherichia coli'' (),Wells, J. C. (2000) Longman Pronunciation Dictionary. Harlow ngland Pearson Education Ltd. also known as ''E. coli'' (), is a Gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus '' Esc ...
can interact with either of two periplasmic binding proteins and mediates the high affinity uptake of sulfate and thiosulfate. May also be involved in the uptake of selenite, selenate and possibly molybdate
In chemistry a molybdate is a compound containing an oxoanion with molybdenum in its highest oxidation state of 6. Molybdenum can form a very large range of such oxoanions which can be discrete structures or polymeric extended structures, altho ...
. Does not undergo phosphorylation during the transport.
:* ATP + H2O + sulfate ulfate - binding protein ide 1= ADP + phosphate + sulfate ide 2+ ulfate - binding proteinside 1]
7.3.2.4
ABC-type nitrate transporter: The expected taxonomic range for this enzyme is: Eukaryota, Bacteria. The enzyme, found in bacteria, interacts with an extracytoplasmic substrate binding protein and mediates the import of nitrate, nitrite, and cyanate. :* ATP + H2O + nitrate itrate - binding proteinide 1= ADP + phosphate + nitrate ide 2+ itrate - binding proteinside 1]
7.3.2.5
ABC-type molybdate transporter: The expected taxonomic range for this enzyme is: Archaea, Eukaryota, Bacteria. The enzyme, found in bacteria, interacts with an extracytoplasmic substrate binding protein and mediates the high-affinity import of molybdate and
tungstate
In chemistry, a tungstate is a compound that contains an oxyanion of tungsten or is a mixed oxide containing tungsten. The simplest tungstate ion is , "orthotungstate". Many other tungstates belong to a large group of polyatomic ions that are ...
. Does not undergo phosphorylation during the transport process.
:* ATP + H2O + molybdate olybdate - binding proteinide 1= ADP + phosphate + molybdate ide 2+ olybdate - binding proteinside 1]
7.3.2.6
ABC-type tungstate transporter: The expected taxonomic range for this enzyme is: Archaea, Bacteria. The enzyme, characterized from the archaeon
Pyrococcus furiosus
''Pyrococcus furiosus'' is a heterotrophic, strictly anaerobic, extremophilic, model species of archaea. It is classified as a hyperthermophile because it thrives best under extremely high temperatures, and is notable for having an optimum gr ...
, the Gram-positive
In bacteriology, gram-positive bacteria are bacteria that give a positive result in the Gram stain test, which is traditionally used to quickly classify bacteria into two broad categories according to their type of cell wall.
Gram-positive bact ...
bacterium Eubacterium
''Eubacterium'' is a genus of Gram-positive bacteria in the family Eubacteriaceae. These bacteria are characterised by a rigid cell wall
A cell wall is a structural layer surrounding some types of cells, just outside the cell membrane. It ...
acidaminophilum and the Gram-negative
Gram-negative bacteria are bacteria that do not retain the crystal violet stain used in the Gram staining method of bacterial differentiation. They are characterized by their cell envelopes, which are composed of a thin peptidoglycan cell wa ...
bacterium Campylobacter jejuni, interacts with an extracytoplasmic substrate binding protein and mediates the import of tungstate into the cell for incorporation into tungsten-dependent enzymes.
:*ATP + H2O + tungstate ungstate - binding proteinide 1= ADP + phosphate + tungstate ide 2+ ungstate - binding proteinside 1]
EC 7.4 Catalysing the translocation of amino acids and peptides
Subclasses are based on the reaction processes that provide the driving force for the translocation. At present there is only one subclass: E7.4.2
Translocation of amino acids and peptides linked to the hydrolysis of a nucleoside triphosphate.
7.4.2.1
ABC-type polar-amino-acid transporter: The expected taxonomic range for this enzyme is: Eukaryota, Bacteria. The enzyme, found in bacteria, interacts with an extracytoplasmic substrate binding protein and mediates the import of polar amino acids. This entry comprises bacterial enzymes that import
Histidine
Histidine (symbol His or H) is an essential amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated –NH3+ form under biological conditions), a carboxylic acid group (which is in the ...
, Arginine
Arginine is the amino acid with the formula (H2N)(HN)CN(H)(CH2)3CH(NH2)CO2H. The molecule features a guanidino group appended to a standard amino acid framework. At physiological pH, the carboxylic acid is deprotonated (−CO2−) and both the am ...
, Lysine
Lysine (symbol Lys or K) is an α-amino acid that is a precursor to many proteins. It contains an α-amino group (which is in the protonated form under biological conditions), an α-carboxylic acid group (which is in the deprotonated − ...
, Glutamine
Glutamine (symbol Gln or Q) is an α-amino acid that is used in the biosynthesis of proteins. Its side chain is similar to that of glutamic acid, except the carboxylic acid group is replaced by an amide. It is classified as a charge-neutral ...
, Glutamate
Glutamic acid (symbol Glu or E; the ionic form is known as glutamate) is an α-amino acid that is used by almost all living beings in the biosynthesis of proteins. It is a non-essential nutrient for humans, meaning that the human body can synt ...
, Aspartate
Aspartic acid (symbol Asp or D; the ionic form is known as aspartate), is an α-amino acid that is used in the biosynthesis of proteins. Like all other amino acids, it contains an amino group and a carboxylic acid. Its α-amino group is in the pro ...
, ornithine
Ornithine is a non-proteinogenic amino acid that plays a role in the urea cycle. Ornithine is abnormally accumulated in the body in ornithine transcarbamylase deficiency. The radical is ornithyl.
Role in urea cycle
L-Ornithine is one of the produ ...
, octopine
Octopine is a derivative of the amino acids arginine and alanine. It was the first member of the class of chemical compounds known as opines to be discovered. Octopine gets its name from '' Octopus octopodia'' from which it was first isolated ...
and nopaline
Nopaline is a chemical compound derived from the amino acids glutamic acid and arginine. It is classified as an opine. Ti plasmids are classified on the basis of the different types of opines they produce. These may be nopaline plasmids, octopi ...
.
:* ATP + H2O + polar amino acid olar amino acid-binding proteinide 1= ADP + phosphate + polar amino acid ide 2+ olar amino acid-binding proteinside1]
7.4.2.2
ABC-type nonpolar-amino-acid transporter: The expected taxonomic range for this enzyme is: Eukaryota, Bacteria. The enzyme, found in bacteria, interacts with an extracytoplasmic substrate binding protein. This entry comprises enzymes that import
Leucine
Leucine (symbol Leu or L) is an essential amino acid that is used in the biosynthesis of proteins. Leucine is an α-amino acid, meaning it contains an α- amino group (which is in the protonated −NH3+ form under biological conditions), an α- ...
, Isoleucie and Valine
Valine (symbol Val or V) is an α-amino acid that is used in the biosynthesis of proteins. It contains an α- amino group (which is in the protonated −NH3+ form under biological conditions), an α- carboxylic acid group (which is in the deprotona ...
.
:* ATP + H2O + non polar amino acid on polar amino acid - binding proteinide 1= ADP + phosphate + non polar amino acid ide 2+ on polar amino acid - binding proteinside 1]
7.4.2.3
ABC-type mitochondrial protein-transporting ATPase: The expected taxonomic range for this enzyme is: Eukaryota, Bacteria. A non-phosphorylated, non-ABC (ATP-binding cassette) ATPase involved in the transport of proteins or preproteins into mitochondria using the TIM ( Translocase of the inner membrane, Translocase of the Inner Membrane) protein complex. TIM is the protein transport machinery of the mitochondrial inner membrane that contains three essential TIM proteins: Tim17 and Tim23 are thought to build a preprotein translocation channel while Tim44 interacts transiently with the matrix heat-shock protein
Hsp70
The 70 kilodalton heat shock proteins (Hsp70s or DnaK) are a family of conserved ubiquitously expressed heat shock proteins. Proteins with similar structure exist in virtually all living organisms. Intracellularly localized Hsp70s are an import ...
to form an ATP-driven import motor.
:* ATP + H2O + mitochondrial protein ide 1= ADP + phosphate + mitochondrial protein ide 2
7.4.2.4
ABC-type chloroplast protein-transporting ATPase: The enzyme appears in viruses and cellular organisms. Involved in the transport of proteins or preproteins into chloroplast stroma (several ATPases may participate in this process). :* ATP + H2O + chloroplast protein ide 1= ADP + phosphate + chloroplast protein ide 2
7.4.2.5
ABC-type protein transporter: The expected taxonomic range for this enzyme is: Eukaryota, Bacteria. This entry stands for a family of bacterial enzymes that are dedicated to the secretion of one or several closely related proteins belonging to the toxin,
protease
A protease (also called a peptidase, proteinase, or proteolytic enzyme) is an enzyme that catalyzes (increases reaction rate or "speeds up") proteolysis, breaking down proteins into smaller polypeptides or single amino acids, and spurring the ...
and lipase
Lipase ( ) is a family of enzymes that catalyzes the hydrolysis of fats. Some lipases display broad substrate scope including esters of cholesterol, phospholipids, and of lipid-soluble vitamins and sphingomyelinases; however, these are usually ...
families. Examples from Gram-negative bacteria include α-hemolysin, cyclolysin, colicin V and siderophores, while examples from Gram-positive bacteria include bacteriocin, subtilin, competence factor and pediocin.
:* ATP + H2O + protein ide 1= ADP + phosphate + protein ide 2
7.4.2.6
ABC-type oligopeptide transporter: A bacterial enzyme that interacts with an extracytoplasmic substrate binding protein and mediates the import of
oligopeptides
An oligopeptide, often just called peptide ('' oligo-'', "a few"), consists of two to twenty amino acids and can include dipeptides, tripeptides, tetrapeptides, and pentapeptides. Some of the major classes of naturally occurring oligopeptides inc ...
of varying nature. The binding protein determines the specificity of the system. Does not undergo phosphorylation during the transport process.
:* ATP + H2O + oligopeptide ligopeptide - binding proteinide 1= ADP + phosphate + oligopeptide ide 2+ ligopeptide - binding proteinside 1]
7.4.2.7
ABC-type alpha-factor-pheromone transporter: The enzyme appears in viruses and cellular organisms characterized by the presence of two similar ATP-binding domains/proteins and two integral membrane domains/proteins. Does not undergo phosphorylation during the transport process. A yeast enzyme that exports the α-factor sex pheromone. :* ATP + H2O + alpha factor ide 1= ADP + phosphate + alpha factor ide 2
7.4.2.8
ABC-type protein-secreting ATPase: The expected taxonomic range for this enzyme is: Archaea, Bacteria. A non-phosphorylated, non-ABC (ATP-binding cassette) ATPase that is involved in protein transport. :* ATP + H2O + cellular protein ide 1= ADP + phosphate + cellular protein ide 2
7.4.2.9
ABC-type dipeptide transporter: The enzyme appears in viruses and cellular organisms. ATP-binding cassette (ABC) type transporter, characterized by the presence of two similar ATP-binding domains/proteins and two integral membrane domains/proteins. A bacterial enzyme that interacts with an extracytoplasmic substrate binding protein and mediates the uptake of dipeptides and tripeptides. :* ATP + H2O + dipeptide ipeptide - binding proteinide 1= ADP + phosphate + ide 2+ ipeptide - binding proteinside 1]
7.4.2.10
ABC-type glutathione transporter: A prokaryotic ATP-binding cassette (ABC) type transporter, characterized by the presence of two similar ATP-binding domains/proteins and two integral membrane domains/proteins. The enzyme from the bacterium ''
Escherichia coli
''Escherichia coli'' (),Wells, J. C. (2000) Longman Pronunciation Dictionary. Harlow ngland Pearson Education Ltd. also known as ''E. coli'' (), is a Gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus '' Esc ...
'' is a heterotrimeric complex that interacts with an extracytoplasmic substrate binding protein to mediate the uptake of glutathione
Glutathione (GSH, ) is an antioxidant in plants, animals, fungi, and some bacteria and archaea. Glutathione is capable of preventing damage to important cellular components caused by sources such as reactive oxygen species, free radicals, pe ...
.
:* ATP + H2O glutathione lutathione - binding proteinide 1= ADP + phosphate + glutathione ide 2+ lutathione - binding proteinside 1]
7.4.2.11
ABC-type methionine transporter: A bacterial enzyme that interacts with an extracytoplasmic substrate binding protein and functions to import
methionine
Methionine (symbol Met or M) () is an essential amino acid in humans. As the precursor of other amino acids such as cysteine and taurine, versatile compounds such as SAM-e, and the important antioxidant glutathione, methionine plays a critical ...
.
:* (1) ATP + H2O + L-methionine ethionine - binding proteinide 1= ADP + phosphate + L-methionine ide 2+ ethionine - binding proteinside 1]
:* (2) ATP + H2O + D-methionine ethionine - binding proteinide 1= ADP + phosphate + D-methionine ide 2+ ethionine - binding proteinside 1]
7.4.2.12
ABC-type cystine transporter: A bacterial enzyme that interacts with an extracytoplasmic substrate binding protein and mediates the high affinity import of trace cystine. The enzyme from ''
Escherichia coli
''Escherichia coli'' (),Wells, J. C. (2000) Longman Pronunciation Dictionary. Harlow ngland Pearson Education Ltd. also known as ''E. coli'' (), is a Gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus '' Esc ...
'' K-12 can import both isomers of cystine and a variety of related molecules including djenkolate, lanthionine, diaminopimelate and homocystine
Homocystine is a chemical compound consisting of two homocysteine molecules joined by a disulfide bond. Its relationship with homocysteine is analogous to the relationship between cystine and cysteine.
References
{{organic-compound-stub
...
.
:* (1) ATP + H2O + L-cystine ystine - binding proteinide 1= ADP + phosphate + L-cystine ide 2+ ystine - binding proteinside 1]
:* (2) ATP + H2O + D-cystine ystine - binding proteinide 1= ADP + phosphate + D-cystine ide 2+ ystine - binding proteinside 1]
EC 7.5 Catalysing the translocation of carbohydrates and their derivatives
EC 7.5.2
Linked to the hydrolysis of a
nucleoside triphosphate
A nucleoside triphosphate is a nucleoside containing a nitrogenous base bound to a 5-carbon sugar (either ribose or deoxyribose), with three phosphate groups bound to the sugar. They are the molecular precursors of both DNA and RNA, which ar ...
EC 7.6 Catalysing the translocation of other compounds
EC 7.6.2
Linked to the hydrolysis of a
nucleoside triphosphate
A nucleoside triphosphate is a nucleoside containing a nitrogenous base bound to a 5-carbon sugar (either ribose or deoxyribose), with three phosphate groups bound to the sugar. They are the molecular precursors of both DNA and RNA, which ar ...
Examples
* ornithine translocase (SLC25A15), associated with ornithine translocase deficiency. *carnitine-acylcarnitine translocase
Carnitine-acylcarnitine translocase (CACT) is responsible for passive transport of carnitine and carnitine- fatty acid complexes and across the inner mitochondrial membrane as part of the carnitine shuttle system.
Function
Fatty acyl–carnitin ...
(SLC25A20), associated with carnitine-acylcarnitine translocase deficiency
Carnitine-acylcarnitine translocase deficiency is a rare, autosomal recessive metabolic disorder that prevents the body from converting long-chain fatty acids into energy, particularly during periods without food. Carnitine, a natural substance ac ...
.
* Translocase of outer mitochondrial membrane 40 ( TOMM40), a protein encoded by the TOMM40 gene, whose alleles differentially impact the risk for Alzheimer's disease
Alzheimer's disease (AD) is a neurodegenerative disease that usually starts slowly and progressively worsens. It is the cause of 60–70% of cases of dementia. The most common early symptom is difficulty in remembering recent events. As ...
References
{{Portal bar, Biology, border=no Translocases Membrane proteins Solute carrier family