Systematic Evolution of Ligands by Exponential Enrichment on:
[Wikipedia]
[Google]
[Amazon]
 Systematic evolution of ligands by exponential enrichment (SELEX), also referred to as '' in vitro selection'' or '' in vitro evolution'', is a
Systematic evolution of ligands by exponential enrichment (SELEX), also referred to as '' in vitro selection'' or '' in vitro evolution'', is a
NeoVentures Biotechnology Inc
to allow the selection of aptamers without immobilizing the target or the oligonucleotide library. Immobilization is a necessary component of SELEX; however, it has the potential to inhibit key epitopes, and thus weaken the likelihood of successful binding, particularly when working with small molecules. FRELEX follows a similar overall methodology to SELEX; however, instead of immobilizing the target, the researcher introduces a series of random and blocker oligonucleotides to an immobilization field before introduction to the target. This allows the researcher to better target small molecules that may be lost during partitioning. It also can be used in some circumstances to select an aptamer library without knowing the target.
Aptamer Base
Evolution Genetics techniques Molecular biology
 Systematic evolution of ligands by exponential enrichment (SELEX), also referred to as '' in vitro selection'' or '' in vitro evolution'', is a
Systematic evolution of ligands by exponential enrichment (SELEX), also referred to as '' in vitro selection'' or '' in vitro evolution'', is a combinatorial chemistry Combinatorial chemistry comprises chemical synthetic methods that make it possible to prepare a large number (tens to thousands or even millions) of compounds in a single process. These compound libraries can be made as mixtures, sets of individua ...
technique in molecular biology
Molecular biology is the branch of biology that seeks to understand the molecular basis of biological activity in and between cells, including biomolecular synthesis, modification, mechanisms, and interactions. The study of chemical and physi ...
for producing oligonucleotide
Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, research, and forensics. Commonly made in the laboratory by solid-phase chemical synthesis, these small bits of nucleic acids ...
s of either single-stranded DNA or RNA that specifically bind to a target ligand
In coordination chemistry, a ligand is an ion or molecule ( functional group) that binds to a central metal atom to form a coordination complex. The bonding with the metal generally involves formal donation of one or more of the ligand's elec ...
or ligands. These single-stranded DNA or RNA are commonly referred to as aptamer
Aptamers are short sequences of artificial DNA, RNA, XNA, or peptide that bind a specific target molecule, or family of target molecules. They exhibit a range of affinities ( KD in the pM to μM range), with little or no off-target bindin ...
s.
Although SELEX has emerged as the most commonly used name for the procedure, some researchers have referred to it as SAAB (selected and amplified binding site) and CASTing (cyclic amplification and selection of targets) SELEX was first introduced in 1990. In 2015, a special issue was published in the Journal of Molecular Evolution in the honor of quarter century of the discovery of SELEX.
The process begins with the synthesis of a very large oligonucleotide library, consisting of randomly generated sequences of fixed length flanked by constant 5' and 3' ends. The constant ends serve as primers, while a small number of random regions are expected to bind specifically to the chosen target. For a randomly generated region of length ''n'', the number of possible sequences in the library using conventional DNA or RNA is 4n (''n'' positions with four possibilities (A,T,C,G) at each position). The sequences in the library are exposed to the target ligand - which may be a protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, res ...
or a small organic compound - and those that do not bind the target are removed, usually by affinity chromatography
Affinity chromatography is a method of separating a biomolecule from a mixture, based on a highly specific macromolecular binding interaction between the biomolecule and another substance. The specific type of binding interaction depends on the ...
or target capture on paramagnetic beads. The bound sequences are eluted and amplified by PCR to prepare for subsequent rounds of selection in which the stringency of the elution conditions can be increased to identify the tightest-binding sequences. A caution to consider in this method is that the selection of extremely high, sub- nanomolar binding affinity entities may not in fact improve specificity for the target molecule. Off-target binding to related molecules could have significant clinical effects.
SELEX has been used to develop a number of aptamers that bind targets interesting for both clinical and research purposes. Nucleotides with chemically modified sugars and bases have been incorporated into SELEX reactions to increase the chemical diversity at each base, expanding the possibilities for specific and sensitive binding, or increasing stability in serum or ''in vivo
Studies that are ''in vivo'' (Latin for "within the living"; often not italicized in English) are those in which the effects of various biological entities are tested on whole, living organisms or cells, usually animals, including humans, and ...
''.
Procedure
Aptamers have emerged as a novel category in the field of bioreceptors due to their wide applications ranging from biosensing to therapeutics. Several variations of their screening process, called SELEX have been reported which can yield sequences with desired properties needed for their final use.Generating single stranded oligonucleotide library
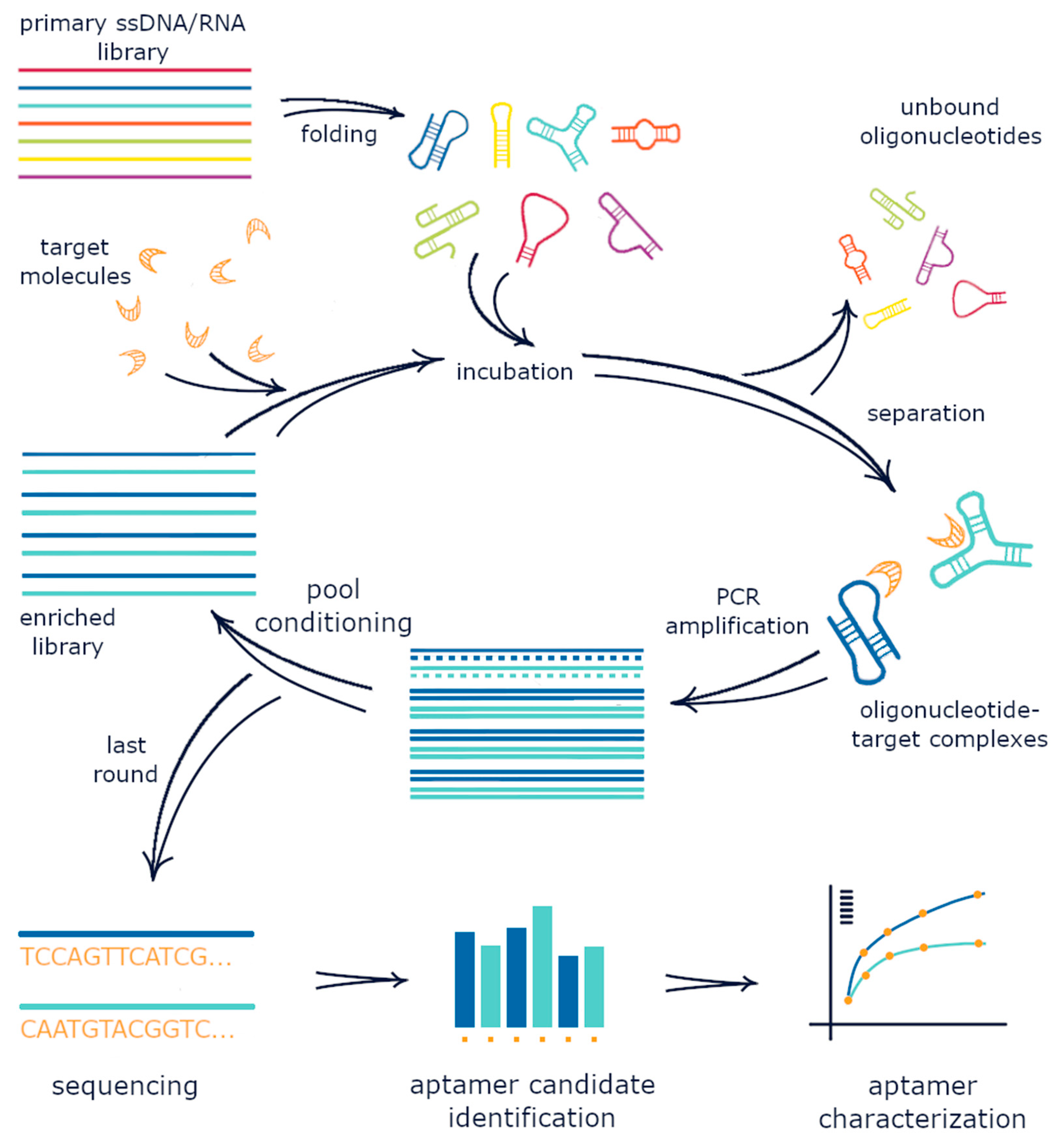
The first step of SELEX involves the synthesis of fully or partially randomized oligonucleotide sequences of some length flanked by defined regions which allow PCR amplification of those randomized regions and, in the case of RNA SELEX, in vitro transcription of the randomized sequence. While Ellington and Szostak demonstrated that chemical synthesis is capable of generating ~1015 unique sequences for oligonucleotide libraries in their 1990 paper on in vitro selection, they found that amplification of these synthesized oligonucleotides led to significant loss of pool diversity due to PCR bias and defects in synthesized fragments. The oligonucleotide pool is amplified and a sufficient amount of the initial library is added to the reaction so that there are numerous copies of each individual sequence to minimize the loss of potential binding sequences due to stochastic events. Before the library is introduced to target for incubation and selective retention, the sequence library must be converted to single stranded oligonucleotides to achieve structural conformations with target binding properties.Target incubation
Immediately prior to target introduction, the single stranded oligonucleotide library is often heated and cooled slowly to renature oligonucleotides into thermodynamically stable secondary and tertiary structures. Once prepared, the randomized library is incubated with immobilized target to allow oligonucleotide-target binding. There are several considerations for this target incubation step, including the target immobilization method and strategies for subsequent unbound oligonucleotide separation, incubation time and temperature, incubation buffer conditions, and target versus oligonucleotide concentrations. Examples of target immobilization methods includeaffinity chromatography
Affinity chromatography is a method of separating a biomolecule from a mixture, based on a highly specific macromolecular binding interaction between the biomolecule and another substance. The specific type of binding interaction depends on the ...
columns, nitrocellulose binding assay filters, and paramagnetic beads. Recently, SELEX reactions have been developed where the target is whole cells, which are expanded near complete confluence and incubated with the oligonucleotide library on culture plates. Incubation buffer conditions are altered based on the intended target and desired function of the selected aptamer. For example, in the case of negatively charged small molecules and proteins, high salt buffers are used for charge screening to allow nucleotides to approach the target and increase the chance of a specific binding event. Alternatively, if the desired aptamer function is in vivo protein or whole cell binding for potential therapeutic or diagnostic application, incubation buffer conditions similar to in vivo plasma salt concentrations and homeostatic temperatures are more likely to generate aptamers that can bind in vivo. Another consideration in incubation buffer conditions is non-specific competitors. If there is a high likelihood of non-specific oligonucleotide retention in the reaction conditions, non specific competitors, which are small molecules or polymers other than the SELEX library that have similar physical properties to the library oligonucleotides, can be used to occupy these non-specific binding sites. Varying the relative concentration of target and oligonucleotides can also affect properties of the selected aptamers. If a good binding affinity
In biochemistry and pharmacology, a ligand is a substance that forms a complex with a biomolecule to serve a biological purpose. The etymology stems from ''ligare'', which means 'to bind'. In protein-ligand binding, the ligand is usually a m ...
for the selected aptamer is not a concern, then an excess of target can be used to increase the probability that at least some sequences will bind during incubation and be retained. However, this provides no selective pressure for high binding affinity
In biochemistry and pharmacology, a ligand is a substance that forms a complex with a biomolecule to serve a biological purpose. The etymology stems from ''ligare'', which means 'to bind'. In protein-ligand binding, the ligand is usually a m ...
, which requires the oligonucleotide library to be in excess so that there is competition between unique sequences for available specific binding sites.
Binding sequence elution and amplification
Once the oligonucleotide library has been incubated with target for sufficient time, unbound oligonucleotides are washed away from immobilized target, often using the incubation buffer so that specifically bound oligonucleotides are retained. With unbound sequences washed away, the specifically bound sequences are then eluted by creating denaturing conditions that promote oligonucleotide unfolding or loss of binding conformation including flowing in deionized water, using denaturing solutions containing urea and EDTA, or by applying high heat and physical force. Upon elution of bound sequences, the retained oligonucleotides are reverse-transcribed to DNA in the case of RNA or modified base selections, or simply collected for amplification in the case of DNA SELEX. These DNA templates from eluted sequences are then amplified via PCR and converted to single stranded DNA, RNA, or modified base oligonucleotides, which are used as the initial input for the next round of selection.Obtaining ssDNA
One of the most critical steps in the SELEX procedure is obtaining single stranded DNA (ssDNA) after the PCR amplification step. This will serve as input for the next cycle so it is of vital importance that all the DNA is single stranded and as little as possible is lost. Because of the relative simplicity, one of the most used methods is using biotinylated reverse primers in the amplification step, after which the complementary strands can be bound to a resin followed by elution of the other strand with lye. Another method is asymmetric PCR, where the amplification step is performed with an excess of forward primer and very little reverse primer, which leads to the production of more of the desired strand. A drawback of this method is that the product should be purified from double stranded DNA (dsDNA) and other left-over material from the PCR reaction. Enzymatic degradation of the unwanted strand can be performed by tagging this strand using a phosphate-probed primer, as it is recognized by enzymes such as Lambda exonuclease. These enzymes then selectively degrade the phosphate tagged strand leaving the complementary strand intact. All of these methods recover approximately 50 to 70% of the DNA. For a detailed comparison refer to the article by Svobodová et al. where these, and other, methods are experimentally compared. In classical SELEX, the process of randomized single stranded library generation, target incubation, and binding sequence elution and amplification described above are repeated until the vast majority of the retained pool consists of target binding sequences, though there are modifications and additions to the procedure that are often used, which are discussed below.Negative or counter selection
In order to increase the specificity of aptamers selected by a given SELEX procedure, a negative selection, or counter selection, step can be added prior to or immediately following target incubation. To eliminate sequences with affinity for target immobilization matrix components from the pool, negative selection can be used where the library is incubated with target immobilization matrix components and unbound sequences are retained. Negative selection can also be used to eliminate sequences that bind target-like molecules or cells by incubating the oligonucleotide library with small molecule target analogs, undesired cell types, or non-target proteins and retaining the unbound sequences.Tracking selection progression
To track the progress of a SELEX reaction, the number of target bound molecules, which is equivalent to the number of oligonucleotides eluted, can be compared to the estimated total input of oligonucleotides following elution at each round. The number of eluted oligonucleotides can be estimated through elution concentration estimations via 260 nm wavelength absorbance or fluorescent labeling of oligonucleotides. As the SELEX reaction approaches completion, the fraction of the oligonucleotide library that binds target approaches 100%, such that the number of eluted molecules approaches the total oligonucleotide input estimate, but may converge at a lower number.Caveats and considerations
Some SELEX reactions can generate probes that are dependent on primer binding regions for secondary structure formation. There are aptamer applications for which a short sequence, and thus primer truncation, is desirable. An advancement on the original method allows an RNA library to omit the constant primer regions, which can be difficult to remove after the selection process because they stabilize secondary structures that are unstable when formed by the random region alone.Chemically modified nucleotides
Recently, SELEX has expanded to include the use of chemically modified nucleotides. These chemically modified oligonucleotides offer many potential advantages for selected aptamers including greater stability and nuclease resistance, enhanced binding for select targets, expanded physical properties - like increased hydrophobicity, and more diverse structural conformations. The genetic alphabet, and thus possible aptamers, is also expanded using unnatural base pairs the use of these unnatural base pairs was applied to SELEX and high affinity DNA aptamers were generated.SELEX variants and alternative aptamer selection methods
FRELEX was developed in 2016 bNeoVentures Biotechnology Inc
to allow the selection of aptamers without immobilizing the target or the oligonucleotide library. Immobilization is a necessary component of SELEX; however, it has the potential to inhibit key epitopes, and thus weaken the likelihood of successful binding, particularly when working with small molecules. FRELEX follows a similar overall methodology to SELEX; however, instead of immobilizing the target, the researcher introduces a series of random and blocker oligonucleotides to an immobilization field before introduction to the target. This allows the researcher to better target small molecules that may be lost during partitioning. It also can be used in some circumstances to select an aptamer library without knowing the target.
Prior targets
The technique has been used to evolveaptamer
Aptamers are short sequences of artificial DNA, RNA, XNA, or peptide that bind a specific target molecule, or family of target molecules. They exhibit a range of affinities ( KD in the pM to μM range), with little or no off-target bindin ...
s of extremely high binding affinity to a variety of target ligands, including small molecule
Within the fields of molecular biology and pharmacology, a small molecule or micromolecule is a low molecular weight (≤ 1000 daltons) organic compound that may regulate a biological process, with a size on the order of 1 nm. Many drugs ...
s such as ATP and adenosine and proteins such as prions and vascular endothelial growth factor
Vascular endothelial growth factor (VEGF, ), originally known as vascular permeability factor (VPF), is a signal protein produced by many cells that stimulates the formation of blood vessels. To be specific, VEGF is a sub-family of growth factors, ...
(VEGF). Moreover, SELEX has been used to select high-affinity aptamers for complex targets such as tumor cells, tumor exosomes, or tumor tissue. Clinical uses of the technique are suggested by aptamers that bind tumor marker
A tumor marker is a biomarker found in blood, urine, or body tissues that can be elevated by the presence of one or more types of cancer. There are many different tumor markers, each indicative of a particular disease process, and they are used in ...
s, GFP-related fluorophores
A fluorophore (or fluorochrome, similarly to a chromophore) is a fluorescent chemical compound that can re-emit light upon light excitation. Fluorophores typically contain several combined aromatic groups, or planar or cyclic molecules with sev ...
, and a VEGF-binding aptamer trade-named Macugen
Pegaptanib sodium injection (brand name Macugen) is an anti-angiogenic medicine for the treatment of neovascular (wet) age-related macular degeneration (AMD). It was discovered by NeXstar Pharmaceuticals (which merged with Gilead Sciences in 199 ...
has been approved by the FDA for treatment of macular degeneration
Macular degeneration, also known as age-related macular degeneration (AMD or ARMD), is a medical condition which may result in blurred or no vision in the center of the visual field. Early on there are often no symptoms. Over time, however, som ...
. Additionally, SELEX has been utilized to obtain highly specific catalytic DNA or DNAzymes. Several metal-specific DNAzymes have been reported including the GR-5 DNAzyme (lead-specific), the CA1-3 DNAzymes (copper-specific), the 39E DNAzyme (uranyl-specific) and the NaA43 DNAzyme (sodium-specific).
These developed aptamers have seen diverse application in therapies for macular degeneration and various research applications including biosensor
A biosensor is an analytical device, used for the detection of a chemical substance, that combines a biological component with a physicochemical detector.
The ''sensitive biological element'', e.g. tissue, microorganisms, organelles, cell rece ...
s, fluorescent labeling of proteins and cells, and selective enzyme inhibition.
See also
* * * *References
Further reading
* * {{refendExternal links
Aptamer Base
Evolution Genetics techniques Molecular biology