Somatic Hypermutation on:
[Wikipedia]
[Google]
[Amazon]
Somatic hypermutation (or SHM) is a cellular mechanism by which the immune system adapts to the new foreign elements that confront it (e.g. microbes), as seen during
''Antibody Repertoires and Pathogen Recognition:''
The Role of Germline Diversity and Somatic Hypermutation'' (Thesis) University of Leeds.'' Because this mechanism is merely selective and not precisely targeted, somatic hypermutation has been strongly implicated in the development of
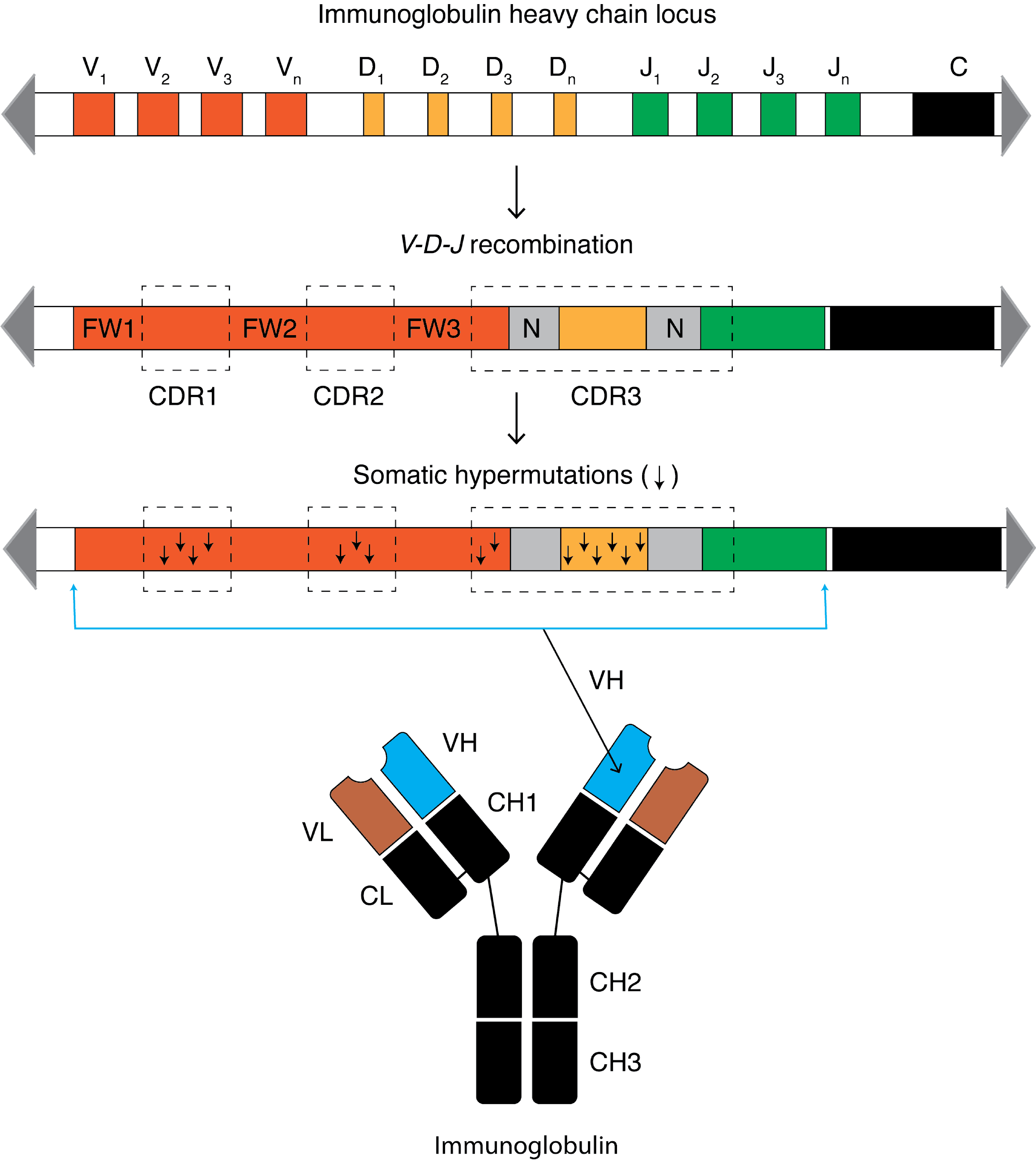 When a B cell recognizes an antigen, it is stimulated to divide (or proliferate). During proliferation, the
When a B cell recognizes an antigen, it is stimulated to divide (or proliferate). During proliferation, the

 The mechanism of SHM involves deamination of
The mechanism of SHM involves deamination of

 The more controversial competing mechanism is an RNA/RT-based mechanism (reverse transcriptase model of SHM) which attempts to explain the production of the full spectrum of strand-biased mutations at A:T and G:C base pairs whereby mutations of A are observed to exceed mutations of T (A>>>T) and mutations of G are observed to exceed mutations of C (G>>>C). This involves error-prone cDNA synthesis via an RNA-dependent DNA polymerase copying the base modified Ig pre-mRNA template and integrating the now error-filled cDNA copy back into the normal chromosomal site. The errors in the Ig pre-mRNA are a combination of adenosine-to-inosine (A-to-I) RNA editing and RNA polymerase II transcription elongation complex copying uracil and abasic sites (arising as AID-mediated lesions) into the nascent pre-mRNA using the transcribed (TS) DNA as the copying template strand. The modern form of this mechanism thus critically depends on AID C-to-U DNA lesions and long tract error-prone cDNA synthesis of the transcribed strand by DNA polymerase-eta acting as a reverse transcriptase.
The evidence for and against each mechanism is critically evaluated in Steele showing that all the molecular data on SHM published since 1980 supports directly or indirectly this RNA/RT-based mechanism.
Recently Zheng et al. have supplied critical independent validation by showing that Adenosine Deaminase enzymes acting on RNA (ADARs) can A-to-I edit both the RNA and DNA moieties of RNA:DNA hybrids in biochemical assays in vitro. RNA:DNA hybrids of about 11 nucleotides in length are transient structures formed at transcription bubbles in vivo during RNA polymerase II elongation.
A preliminary analysis of the implications of the Zheng et al. data has been submitted as formal paper to a refereed journal by Steele and Lindley. The Zheng et al. data strongly imply that the RNA moiety would need to be first A-to-I RNA edited then reverse transcribed and integrated to generate the strong A>>>T strand biased mutation signatures at A:T base pairs observed in all SHM and cancer hypermutation data sets. Editing (A-to-I) of the DNA moiety at RNA:DNA hybrids in vivo cannot explain the A>>T strand bias because such direct DNA modifications would result in T>>>A strand bias which is not observed in any SHM or cancer data set in vivo.
In this regard Robyn Lindley has also recently discovered that the Ig-SHM-like strand-biased mutations in cancer genome protein-coding genes are also in "codon-context". Lindley has termed this process targeted somatic mutation (TSM) to highlight that somatic mutations are far more targeted than previously thought in somatic tissues associated with disease.
The TSM process implies an "in-frame DNA reader" whereby DNA and RNA deaminases at transcribed regions are guided in their mutagenic action, by the codon reading frame of the DNA.
The more controversial competing mechanism is an RNA/RT-based mechanism (reverse transcriptase model of SHM) which attempts to explain the production of the full spectrum of strand-biased mutations at A:T and G:C base pairs whereby mutations of A are observed to exceed mutations of T (A>>>T) and mutations of G are observed to exceed mutations of C (G>>>C). This involves error-prone cDNA synthesis via an RNA-dependent DNA polymerase copying the base modified Ig pre-mRNA template and integrating the now error-filled cDNA copy back into the normal chromosomal site. The errors in the Ig pre-mRNA are a combination of adenosine-to-inosine (A-to-I) RNA editing and RNA polymerase II transcription elongation complex copying uracil and abasic sites (arising as AID-mediated lesions) into the nascent pre-mRNA using the transcribed (TS) DNA as the copying template strand. The modern form of this mechanism thus critically depends on AID C-to-U DNA lesions and long tract error-prone cDNA synthesis of the transcribed strand by DNA polymerase-eta acting as a reverse transcriptase.
The evidence for and against each mechanism is critically evaluated in Steele showing that all the molecular data on SHM published since 1980 supports directly or indirectly this RNA/RT-based mechanism.
Recently Zheng et al. have supplied critical independent validation by showing that Adenosine Deaminase enzymes acting on RNA (ADARs) can A-to-I edit both the RNA and DNA moieties of RNA:DNA hybrids in biochemical assays in vitro. RNA:DNA hybrids of about 11 nucleotides in length are transient structures formed at transcription bubbles in vivo during RNA polymerase II elongation.
A preliminary analysis of the implications of the Zheng et al. data has been submitted as formal paper to a refereed journal by Steele and Lindley. The Zheng et al. data strongly imply that the RNA moiety would need to be first A-to-I RNA edited then reverse transcribed and integrated to generate the strong A>>>T strand biased mutation signatures at A:T base pairs observed in all SHM and cancer hypermutation data sets. Editing (A-to-I) of the DNA moiety at RNA:DNA hybrids in vivo cannot explain the A>>T strand bias because such direct DNA modifications would result in T>>>A strand bias which is not observed in any SHM or cancer data set in vivo.
In this regard Robyn Lindley has also recently discovered that the Ig-SHM-like strand-biased mutations in cancer genome protein-coding genes are also in "codon-context". Lindley has termed this process targeted somatic mutation (TSM) to highlight that somatic mutations are far more targeted than previously thought in somatic tissues associated with disease.
The TSM process implies an "in-frame DNA reader" whereby DNA and RNA deaminases at transcribed regions are guided in their mutagenic action, by the codon reading frame of the DNA.
class switching
Immunoglobulin class switching, also known as isotype switching, isotypic commutation or class-switch recombination (CSR), is a biological mechanism that changes a B cell's production of immunoglobulin from one type to another, such as from the ...
. A major component of the process of affinity maturation, SHM diversifies B cell
B cells, also known as B lymphocytes, are a type of white blood cell of the lymphocyte subtype. They function in the humoral immunity component of the adaptive immune system. B cells produce antibody molecules which may be either secreted or ...
receptors used to recognize foreign elements (antigen
In immunology, an antigen (Ag) is a molecule or molecular structure or any foreign particulate matter or a pollen grain that can bind to a specific antibody or T-cell receptor. The presence of antigens in the body may trigger an immune response. ...
s) and allows the immune system to adapt its response to new threats during the lifetime of an organism. Somatic hypermutation involves a programmed process of mutation affecting the variable regions of immunoglobulin genes. Unlike germline mutation
A germline mutation, or germinal mutation, is any detectable variation within germ cells (cells that, when fully developed, become sperm and ova). Mutations in these cells are the only mutations that can be passed on to offspring, when either a m ...
, SHM affects only an organism's individual immune cells, and the mutations are not transmitted to the organism's offspring.Oprea, M. (1999''Antibody Repertoires and Pathogen Recognition:''
The Role of Germline Diversity and Somatic Hypermutation'' (Thesis) University of Leeds.'' Because this mechanism is merely selective and not precisely targeted, somatic hypermutation has been strongly implicated in the development of
B-cell lymphoma
The B-cell lymphomas are types of lymphoma affecting B cells. Lymphomas are "blood cancers" in the lymph nodes. They develop more frequently in older adults and in immunocompromised individuals.
B-cell lymphomas include both Hodgkin's lymphoma ...
s and many other cancers.
Targeting
 When a B cell recognizes an antigen, it is stimulated to divide (or proliferate). During proliferation, the
When a B cell recognizes an antigen, it is stimulated to divide (or proliferate). During proliferation, the B-cell receptor
The B cell receptor (BCR) is a transmembrane protein on the surface of a B cell. A B cell receptor is composed of a membrane-bound immunoglobulin molecule and a signal transduction moiety. The former forms a type 1 transmembrane receptor protei ...
locus
Locus (plural loci) is Latin for "place". It may refer to:
Entertainment
* Locus (comics), a Marvel Comics mutant villainess, a member of the Mutant Liberation Front
* ''Locus'' (magazine), science fiction and fantasy magazine
** ''Locus Award' ...
undergoes an extremely high rate of somatic
Somatic may refer to:
* Somatic (biology), referring to the cells of the body in contrast to the germ line cells
** Somatic cell, a non-gametic cell in a multicellular organism
* Somatic nervous system, the portion of the vertebrate nervous syst ...
mutation that is at least 105–106 fold greater than the normal rate of mutation across the genome. Variation is mainly in the form of single-base substitutions, with insertions and deletions being less common. These mutations occur mostly at "hotspots" in the DNA, which are concentrated in hypervariable region
A hypervariable region (HVR) is a location within nuclear DNA or the D-loop of mitochondrial DNA in which base pairs of nucleotides repeat (in the case of nuclear DNA) or have substitutions (in the case of mitochondrial DNA). Changes or repeats in ...
s. These regions correspond to the complementarity-determining region
Complementarity-determining regions (CDRs) are part of the variable chains in immunoglobulins (antibodies) and T cell receptors, generated by B-cells and T-cells respectively, where these molecules bind to their specific antigen. A set of CDRs co ...
s; the sites involved in antigen recognition on the immunoglobulin. The "hotspots" of somatic hypermutation vary depending on the base that is being mutated. RGYW for a G, WRCY for a C, WA for an A and TW for a T. The overall result of the hypermutation process is achieved by a balance between error-prone and high fidelity repair. This directed hypermutation allows for the selection of B cells that express immunoglobulin receptors possessing an enhanced ability to recognize and bind a specific foreign antigen
In immunology, an antigen (Ag) is a molecule or molecular structure or any foreign particulate matter or a pollen grain that can bind to a specific antibody or T-cell receptor. The presence of antigens in the body may trigger an immune response. ...
.
Mechanisms
cytosine
Cytosine () (symbol C or Cyt) is one of the four nucleobases found in DNA and RNA, along with adenine, guanine, and thymine (uracil in RNA). It is a pyrimidine derivative, with a heterocyclic aromatic ring and two substituents attached (an amin ...
to uracil in DNA by the enzyme activation-induced cytidine deaminase
Activation-induced cytidine deaminase, also known as AICDA, AID and single-stranded DNA cytosine deaminase, is a 24 kDa enzyme which in humans is encoded by the ''AICDA'' gene. It creates mutations in DNA by deamination of cytosine base, which ...
, or AID. A cytosine:guanine
Guanine () (symbol G or Gua) is one of the four main nucleobases found in the nucleic acids DNA and RNA, the others being adenine, cytosine, and thymine (uracil in RNA). In DNA, guanine is paired with cytosine. The guanine nucleoside is called ...
pair is thus directly mutated to a uracil:guanine mismatch. Uracil residues are not normally found in DNA, therefore, to maintain the integrity of the genome, most of these mutations must be repaired by high-fidelity Base excision repair enzymes. The uracil bases are removed by the repair enzyme, uracil-DNA glycosylase. Error-prone DNA polymerases are then recruited to fill in the gap and create mutations.
The synthesis of this new DNA involves error-prone DNA polymerase
A DNA polymerase is a member of a family of enzymes that catalyze the synthesis of DNA molecules from nucleoside triphosphates, the molecular precursors of DNA. These enzymes are essential for DNA replication and usually work in groups to create ...
s, which often introduce mutations at the position of the deaminated cytosine itself or neighboring base pair
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA ...
s. During B cell division the immunoglobulin variable region DNA is transcribed and translated. The introduction of mutations in the rapidly proliferating population of B cells ultimately culminates in the production of thousands of B cells, possessing slightly different receptors and varying specificity for the antigen, from which the B cell with highest affinities for the antigen can be selected. The B cells with the greatest affinity will then be selected to differentiate into plasma cell
Plasma cells, also called plasma B cells or effector B cells, are white blood cells that originate in the lymphoid organs as B lymphocytes and secrete large quantities of proteins called antibodies in response to being presented specific substa ...
s producing antibody
An antibody (Ab), also known as an immunoglobulin (Ig), is a large, Y-shaped protein used by the immune system to identify and neutralize foreign objects such as pathogenic bacteria and viruses. The antibody recognizes a unique molecule of the ...
and long-lived memory B cell
In immunology, a memory B cell (MBC) is a type of B lymphocyte that forms part of the adaptive immune system. These cells develop within germinal centers of the secondary lymphoid organs. Memory B cells circulate in the blood stream in a quiescen ...
s contributing to enhanced immune responses upon reinfection.
The hypermutation process also utilizes cells that auto-select against the 'signature' of an organism's own cells. It is hypothesized that failures of this auto-selection process may also lead to the development of an auto-immune
In immunology, autoimmunity is the system of immune responses of an organism against its own healthy cells, tissues and other normal body constituents. Any disease resulting from this type of immune response is termed an "autoimmune disease". P ...
response.
Models
Developments on the viability of the two main competing molecular models on the mechanism of somatic hypermutation (SHM) since 1987 have now reached a resolution, particular molecular data published since 2000. Much of this early phase data has been reviewed by Teng and Papavasiliou and additionally outlined by Di Noia and Maul, and the SHM field data reviewed in Steele and additionally outlined in these papers.DNA deamination model
This can be labelled the DNA-based model. It is enzymatically focused solely on DNA substrates. The modern form, outlined in previous sections is the Neuberger "DNA deamination model" based on activation-induced cytidine deaminase (AID) and short-patch error-prone DNA repair by DNA polymerase-eta operating around AID C-to-U lesions This model only partially explains the origins of the full spectrum of somatic mutations at A:T and G:C base pairs observed in SHM in B lymphocytes in vivo during an antigen-driven immune response. It also does not logically explain how strand biased mutations may be generated. A key feature is its critical dependence on the gap-filling error prone DNA repair synthesis properties of DNA polymerase-eta targeting A:T base pairs at AID-mediated C-to-U lesions or ssDNA nicks. This error-prone DNA polymerase is the only known error-prone polymerase involved in SHM in vivo. What is often ignored in these studies is that this Y family DNA polymerase enzyme is also an efficient reverse transcriptase as demonstrated in ''in vitro'' assays.Reverse transcriptase model
See also
* Affinity maturation *Anergy
In immunology, anergy is a lack of reaction by the body's defense mechanisms to foreign substances, and consists of a direct induction of peripheral lymphocyte tolerance. An individual in a state of anergy often indicates that the immune syste ...
* Immune system
* V(D)J recombination
* Immunoglobulin class switching
Immunoglobulin class switching, also known as isotype switching, isotypic commutation or class-switch recombination (CSR), is a biological mechanism that changes a B cell's production of immunoglobulin from one type to another, such as from the ...
References
External links
* {{Immune system Immune system Antibodies