iron-sulfur cluster on:
[Wikipedia]
[Google]
[Amazon]
Iron–sulfur proteins (or iron–sulphur proteins in

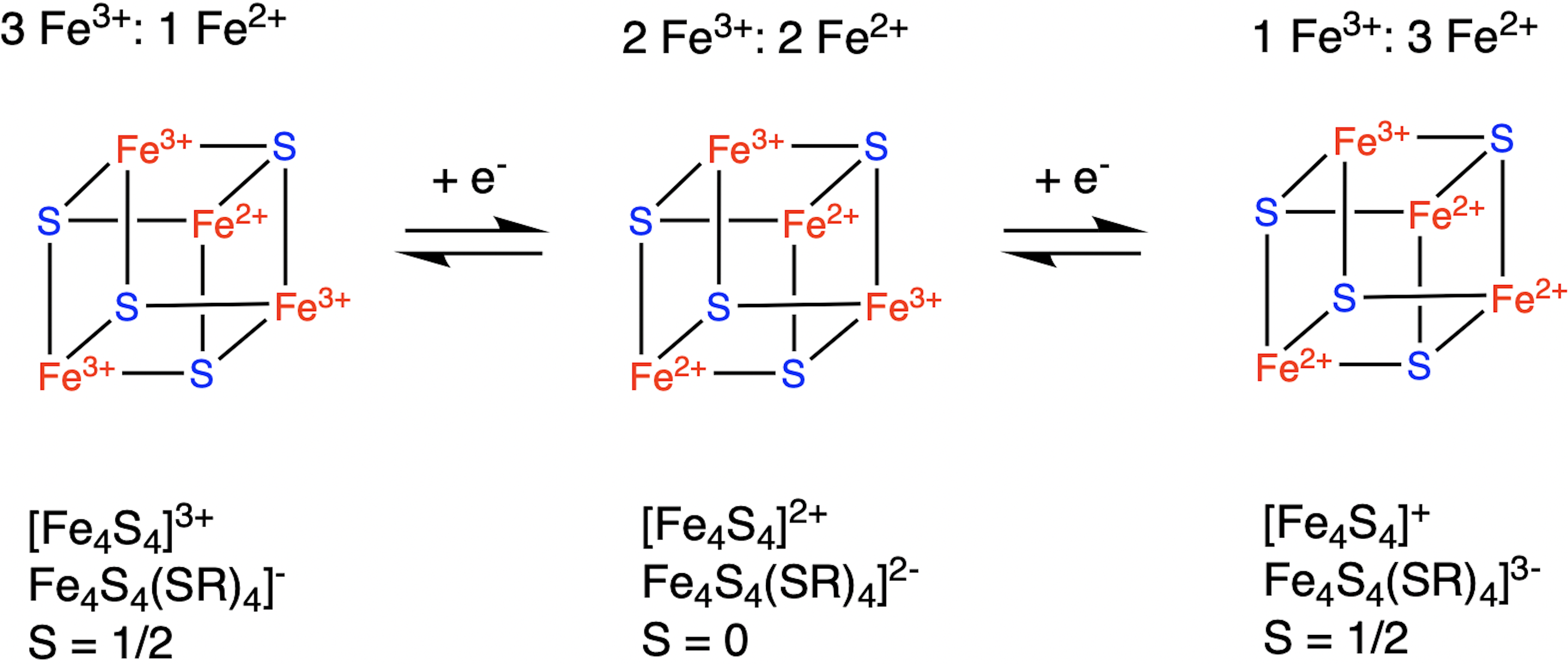 The second cubane shown here with mixed valence pairs (2 Fe3+ and 2 Fe2+), has a greater stability from covalent communication and strong covalent delocalization of the “extra” electron from the reduced Fe2+ that results in full ferromagnetic coupling.
The second cubane shown here with mixed valence pairs (2 Fe3+ and 2 Fe2+), has a greater stability from covalent communication and strong covalent delocalization of the “extra” electron from the reduced Fe2+ that results in full ferromagnetic coupling.

Examples of iron-sulfur clusters
{{DEFAULTSORT:Iron-sulfur protein Cluster chemistry Peripheral membrane proteins Protein structure Iron compounds Sulfur compounds Metalloproteins
British spelling
Despite the various English dialects spoken from country to country and within different regions of the same country, there are only slight regional variations in English orthography, the two most notable variations being British and American ...
) are protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, res ...
s characterized by the presence of iron–sulfur cluster
Iron–sulfur clusters (or iron–sulphur clusters in British spelling) are molecular ensembles of iron and sulfide. They are most often discussed in the context of the biological role for iron–sulfur proteins, which are pervasive. Many Fe– ...
s containing sulfide-linked di-, tri-, and tetrairon centers in variable oxidation state
In chemistry, the oxidation state, or oxidation number, is the hypothetical charge of an atom if all of its bonds to different atoms were fully ionic. It describes the degree of oxidation (loss of electrons) of an atom in a chemical compound. C ...
s. Iron–sulfur clusters are found in a variety of metalloprotein
Metalloprotein is a generic term for a protein that contains a metal ion cofactor. A large proportion of all proteins are part of this category. For instance, at least 1000 human proteins (out of ~20,000) contain zinc-binding protein domains al ...
s, such as the ferredoxins, as well as NADH dehydrogenase
NADH dehydrogenase is an enzyme that converts nicotinamide adenine dinucleotide (NAD) from its reduced form (NADH) to its oxidized form (NAD+). Members of the NADH dehydrogenase family and analogues are commonly systematically named using the for ...
, hydrogenase A hydrogenase is an enzyme that catalyses the reversible oxidation of molecular hydrogen (H2), as shown below:
Hydrogen uptake () is coupled to the reduction of electron acceptors such as oxygen, nitrate, sulfate, carbon dioxide (), and fumarat ...
s, coenzyme Q – cytochrome c reductase
The coenzyme Q : cytochrome ''c'' – oxidoreductase, sometimes called the cytochrome ''bc''1 complex, and at other times complex III, is the third complex in the electron transport chain (), playing a critical role in biochemical generation ...
, succinate – coenzyme Q reductase and nitrogenase
Nitrogenases are enzymes () that are produced by certain bacteria, such as cyanobacteria (blue-green bacteria) and rhizobacteria. These enzymes are responsible for the reduction of nitrogen (N2) to ammonia (NH3). Nitrogenases are the only fa ...
. Iron–sulfur clusters are best known for their role in the oxidation-reduction reactions of electron transport in mitochondria and chloroplasts. Both Complex I and Complex II of oxidative phosphorylation have multiple Fe–S clusters. They have many other functions including catalysis
Catalysis () is the process of increasing the rate of a chemical reaction by adding a substance known as a catalyst (). Catalysts are not consumed in the reaction and remain unchanged after it. If the reaction is rapid and the catalyst recyc ...
as illustrated by aconitase, generation of radicals as illustrated by SAM-dependent enzymes, and as sulfur donors in the biosynthesis of lipoic acid and biotin. Additionally, some Fe–S proteins regulate gene expression. Fe–S proteins are vulnerable to attack by biogenic nitric oxide, forming dinitrosyl iron complexes. In most Fe–S proteins, the terminal ligands on Fe are thiol
In organic chemistry, a thiol (; ), or thiol derivative, is any organosulfur compound of the form , where R represents an alkyl or other organic substituent. The functional group itself is referred to as either a thiol group or a sulfhydryl gro ...
ate, but exceptions exist.
The prevalence of these proteins on the metabolic pathway
In biochemistry, a metabolic pathway is a linked series of chemical reactions occurring within a cell. The reactants, products, and intermediates of an enzymatic reaction are known as metabolites, which are modified by a sequence of chemical reac ...
s of most organisms leads some scientists to theorize that iron–sulfur compounds had a significant role in the origin of life
In biology, abiogenesis (from a- 'not' + Greek bios 'life' + genesis 'origin') or the origin of life is the natural process by which life has arisen from non-living matter, such as simple organic compounds. The prevailing scientific hypothes ...
in the iron–sulfur world theory.
Structural motifs
In almost all Fe–S proteins, the Fe centers are tetrahedral and the terminal ligands are thiolato sulfur centers from cysteinyl residues. The sulfide groups are either two- or three-coordinated. Three distinct kinds of Fe–S clusters with these features are most common.Structure-Function Principles
To serve their various biological roles, iron-sulfur proteins effect rapid electron transfers and span the whole range of physiological redox potentials from -600 mV to +460 mV. Iron-sulfur proteins are involved in various biological electron transport processes, such as photosynthesis and cellular respiration, which require rapid electron transfer to sustain the energy or biochemical needs of the organism. Fe3+-SR bonds have unusually high covalency which is expected. When comparing the covalency of Fe3+ with the covalency of Fe2+, Fe3+ has almost double the covalency of Fe2+ (20% to 38.4%). Fe3+ is also much more stabilized than Fe2+. Hard ions like Fe3+ normally have low covalency because of the energy mismatch of the metal Lowest Unoccupied Molecular Orbital with the ligand Highest Occupied Molecular Orbital. There is HO-H—S-Cys H-bonding from external H2O’s positioned by the protein close to the active site and this H-bonding decreases the lone pair electron donation from the Cys-S donor to the Fe3+/2+. Using lyophilization to remove these external H2O’s results in increased Fe-S covalency, which means that the H2O’s are decreasing the covalency because HOH-S Hydrogen-bonding pulls the sulfur electrons. Since covalency stabilizes Fe3+ more than Fe2+, therefore Fe3+ is more destabilized by the HOH-S hydrogen-bonding. The Fe3+ 3d orbital energies follow the “inverted” bonding scheme which fortuitously has the Fe3+ d-orbitals closely matched in energy with the sulfur 3p orbitals which gives high covalency in the resulting bonding molecular orbital. This high covalency lowers the inner sphere reorganization energy and ultimately contributes to a rapid electron transfer.2Fe–2S clusters
The simplest polymetallic system, the e2S2cluster, is constituted by two iron ions bridged by two sulfide ions and coordinated by fourcysteinyl
Cysteine (symbol Cys or C; ) is a semiessential proteinogenic amino acid with the formula . The thiol side chain in cysteine often participates in enzymatic reactions as a nucleophile.
When present as a deprotonated catalytic residue, some ...
ligand
In coordination chemistry, a ligand is an ion or molecule ( functional group) that binds to a central metal atom to form a coordination complex. The bonding with the metal generally involves formal donation of one or more of the ligand's elec ...
s (in Fe2S2 ferredoxins) or by two cysteines and two histidine
Histidine (symbol His or H) is an essential amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated –NH3+ form under biological conditions), a carboxylic acid group (which is in the d ...
s (in Rieske protein
Rieske proteins are iron–sulfur protein (ISP) components of cytochrome ''bc''1 complexes and cytochrome b6f complexes and are responsible for electron transfer in some biological systems. John S. Rieske and co-workers first discovered the pro ...
s). The oxidized proteins contain two Fe3+ ions, whereas the reduced proteins contain one Fe3+ and one Fe2+ ion. These species exist in two oxidation states, (FeIII)2 and FeIIIFeII. CDGSH iron sulfur domain is also associated with 2Fe-2S clusters.

4Fe–4S clusters
A common motif features a four iron ions and four sulfide ions placed at the vertices of acubane-type cluster
A cubane-type cluster is an arrangement of atoms in a molecular structure that forms a cube. In the idealized case, the eight vertices are symmetry equivalent and the species has Oh symmetry. Such a structure is illustrated by the hydrocarbon ...
. The Fe centers are typically further coordinated by cysteinyl ligands. The e4S4electron-transfer proteins ( e4S4 ferredoxins) may be further subdivided into low-potential (bacterial-type) and high-potential (HiPIP) ferredoxins. Low- and high-potential ferredoxins are related by the following redox scheme:
In HiPIP, the cluster shuttles between Fe3+, 2Fe2+(Fe4S42+) and Fe3+, Fe2+(Fe4S43+). The potentials for this redox couple range from 0.4 to 0.1 V. In the bacterial ferredoxins, the pair of oxidation states are e3+, 3Fe2+(Fe4S4+) and Fe3+, 2Fe2+(Fe4S42+). The potentials for this redox couple range from −0.3 to −0.7 V. The two families of 4Fe–4S clusters share the Fe4S42+ oxidation state. The difference in the redox couples is attributed to the degree of hydrogen bonding, which strongly modifies the basicity of the cysteinyl thiolate ligands. A further redox couple, which is still more reducing than the bacterial ferredoxins is implicated in the nitrogenase
Nitrogenases are enzymes () that are produced by certain bacteria, such as cyanobacteria (blue-green bacteria) and rhizobacteria. These enzymes are responsible for the reduction of nitrogen (N2) to ammonia (NH3). Nitrogenases are the only fa ...
.
Some 4Fe–4S clusters bind substrates and are thus classified as enzyme cofactors. In aconitase, the Fe–S cluster binds aconitate at the one Fe centre that lacks a thiolate ligand. The cluster does not undergo redox, but serves as a Lewis acid catalyst to convert citrate to isocitrate. In radical SAM
Radical SAM is a designation for a superfamily of enzymes that use a +_cluster.html" ;"title="Fe-4Ssup>+ cluster">Fe-4Ssup>+ cluster to reductively cleave ''S''-adenosyl-L-methionine (SAM) to generate a radical, usually a 5′- deoxyadenosyl rad ...
enzymes, the cluster binds and reduces S-adenosylmethionine to generate a radical, which is involved in many biosyntheses.
 The second cubane shown here with mixed valence pairs (2 Fe3+ and 2 Fe2+), has a greater stability from covalent communication and strong covalent delocalization of the “extra” electron from the reduced Fe2+ that results in full ferromagnetic coupling.
The second cubane shown here with mixed valence pairs (2 Fe3+ and 2 Fe2+), has a greater stability from covalent communication and strong covalent delocalization of the “extra” electron from the reduced Fe2+ that results in full ferromagnetic coupling.
3Fe–4S clusters
Proteins are also known to contain e3S4centres, which feature one iron less than the more common e4S4cores. Three sulfide ions bridge two iron ions each, while the fourth sulfide bridges three iron ions. Their formal oxidation states may vary from e3S4sup>+ (all-Fe3+ form) to e3S4sup>2− (all-Fe2+ form). In a number of iron–sulfur proteins, the e4S4cluster can be reversibly converted by oxidation and loss of one iron ion to a e3S4cluster. E.g., the inactive form of aconitase possesses an e3S4and is activated by addition of Fe2+ and reductant.Other Fe–S clusters
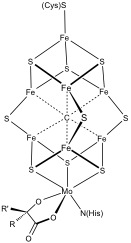
More complex polymetallic systems are common. Examples include both the 8Fe and the 7Fe clusters innitrogenase
Nitrogenases are enzymes () that are produced by certain bacteria, such as cyanobacteria (blue-green bacteria) and rhizobacteria. These enzymes are responsible for the reduction of nitrogen (N2) to ammonia (NH3). Nitrogenases are the only fa ...
. Carbon monoxide dehydrogenase and the eFe
Agencia EFE, S.A. () is a Spanish international news agency, the major multimedia news agency in Spanish language and the world's fourth largest wire service after the Associated Press, Reuters, and Agence France-Presse. EFE was created in 1939 ...
hydrogenase A hydrogenase is an enzyme that catalyses the reversible oxidation of molecular hydrogen (H2), as shown below:
Hydrogen uptake () is coupled to the reduction of electron acceptors such as oxygen, nitrate, sulfate, carbon dioxide (), and fumarat ...
also feature unusual Fe–S clusters. A special 6 cysteine-coordinated e4S3cluster was found in oxygen-tolerant membrane-bound iFehydrogenases.


Biosynthesis
The biosynthesis of the Fe–S clusters has been well studied. The biogenesis of iron sulfur clusters has been studied most extensively in the bacteria '' E. coli'' and '' A. vinelandii'' and yeast '' S. cerevisiae''. At least three different biosynthetic systems have been identified so far, namely nif, suf, and isc systems, which were first identified in bacteria. The nif system is responsible for the clusters in the enzyme nitrogenase. The suf and isc systems are more general. The yeast isc system is the best described. Several proteins constitute the biosynthetic machinery via the isc pathway. The process occurs in two major steps: (1) the Fe/S cluster is assembled on a scaffold protein followed by (2) transfer of the preformed cluster to the recipient proteins. The first step of this process occurs in thecytoplasm
In cell biology, the cytoplasm is all of the material within a eukaryotic cell, enclosed by the cell membrane, except for the cell nucleus. The material inside the nucleus and contained within the nuclear membrane is termed the nucleoplasm. ...
of prokaryotic
A prokaryote () is a single-celled organism that lacks a nucleus and other membrane-bound organelles. The word ''prokaryote'' comes from the Greek πρό (, 'before') and κάρυον (, 'nut' or 'kernel').Campbell, N. "Biology:Concepts & Connec ...
organisms or in the mitochondria of eukaryotic
Eukaryotes () are organisms whose Cell (biology), cells have a cell nucleus, nucleus. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the ...
organisms. In the higher organisms the clusters are therefore transported out of the mitochondrion to be incorporated into the extramitochondrial enzymes. These organisms also possess a set of proteins involved in the Fe/S clusters transport and incorporation processes that are not homologous to proteins found in prokaryotic systems.
Synthetic analogues
Synthetic analogues of the naturally occurring Fe–S clusters were first reported by Holm and coworkers. Treatment of iron salts with a mixture of thiolates and sulfide affords derivatives such as ( Et4N)2Fe4S4(SCH2Ph)4].See also
*Bioinorganic chemistry
Bioinorganic chemistry is a field that examines the role of metals in biology. Bioinorganic chemistry includes the study of both natural phenomena such as the behavior of metalloproteins as well as artificially introduced metals, including those t ...
* Iron-binding proteins Iron-binding proteins are carrier proteins and metalloproteins that are important in iron metabolism and the immune response. Iron is required for life.
Iron-dependent enzymes catalyze a variety of biochemical reactions and can be divided into th ...
* Mitosome
A mitosome is an organelle found in some unicellular eukaryotic organisms, like in members of the supergroup Excavata. The mitosome was found and named in 1999, and its function has not yet been well characterized. It was termed a ''crypton'' by ...
References
*Further reading
* * * * * *External links
*Examples of iron-sulfur clusters
{{DEFAULTSORT:Iron-sulfur protein Cluster chemistry Peripheral membrane proteins Protein structure Iron compounds Sulfur compounds Metalloproteins