Histone-modifying enzymes on:
[Wikipedia]
[Google]
[Amazon]
 Histone-modifying enzymes are
Histone-modifying enzymes are

 Histone phosphorylation occurs when a phosphoryl group is added to a
Histone phosphorylation occurs when a phosphoryl group is added to a
 The presence of O-GlcNAcylation (O-GlcNAc) on serine (S) and threonine (T) histone residues is known to mediate other post-transcriptional histone modifications. The addition and removal of GlcNAc groups are performed by O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA) respectively. While our understanding of these processes is limited, GlcNAcylation of S112 on core histone H2B has been found to promote monoubiquitination of K120. Similarly, OGT associates with the HCF1 complex which interacts with
The presence of O-GlcNAcylation (O-GlcNAc) on serine (S) and threonine (T) histone residues is known to mediate other post-transcriptional histone modifications. The addition and removal of GlcNAc groups are performed by O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA) respectively. While our understanding of these processes is limited, GlcNAcylation of S112 on core histone H2B has been found to promote monoubiquitination of K120. Similarly, OGT associates with the HCF1 complex which interacts with
 ADP-ribosylation (ADPr) defines the addition of one or more adenosine diphosphate ribose (ADP-ribose) groups to a
ADP-ribosylation (ADPr) defines the addition of one or more adenosine diphosphate ribose (ADP-ribose) groups to a



enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products ...
s involved in the modification
Modification may refer to:
* Modifications of school work for students with special educational needs
* Modifications (genetics), changes in appearance arising from changes in the environment
* Posttranslational modifications, changes to prote ...
of histone
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei. They act as spools around which DNA winds to create structural units called nucleosomes. Nucleosomes in turn a ...
substrates after protein translation
In molecular biology and genetics, translation is the process in which ribosomes in the cytoplasm or endoplasmic reticulum synthesize proteins after the process of transcription of DNA to RNA in the cell's nucleus. The entire process ...
and affect cellular processes including gene expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, protein or non-coding RNA, and ultimately affect a phenotype, as the final effect. T ...
. To safely store the eukaryotic
Eukaryotes () are organisms whose cells have a nucleus. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the three domains of life. Bacte ...
genome
In the fields of molecular biology and genetics, a genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding ...
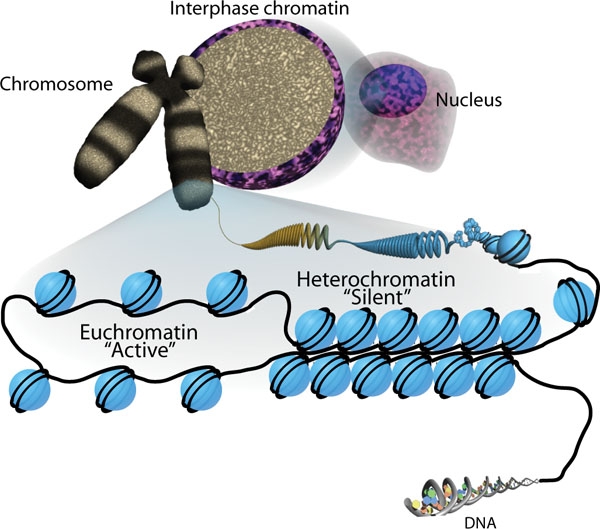
, DNA is wrapped around four core histone
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei. They act as spools around which DNA winds to create structural units called nucleosomes. Nucleosomes in turn a ...
protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, res ...
s (H3, H4, H2A, H2B), which then join to form nucleosome
A nucleosome is the basic structural unit of DNA packaging in eukaryotes. The structure of a nucleosome consists of a segment of DNA wound around eight histone proteins and resembles thread wrapped around a spool. The nucleosome is the fundame ...
s. These nucleosomes further fold together into highly condensed chromatin
Chromatin is a complex of DNA and protein found in eukaryote, eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important ...
, which renders the organism's genetic material far less accessible to the factors required for gene transcription
Transcription is the process of copying a segment of DNA into RNA. The segments of DNA transcribed into RNA molecules that can encode proteins are said to produce messenger RNA (mRNA). Other segments of DNA are copied into RNA molecules called ...
, DNA replication
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all living organisms acting as the most essential part for biological inheritan ...
, recombination and repair
The technical meaning of maintenance involves functional checks, servicing, repairing or replacing of necessary devices, equipment, machinery, building infrastructure, and supporting utilities in industrial, business, and residential installa ...
. Subsequently, eukaryotic organisms have developed intricate mechanisms to overcome this repressive barrier imposed by the chromatin through histone modification, a type of post-translational modification
Post-translational modification (PTM) is the covalent and generally enzymatic modification of proteins following protein biosynthesis. This process occurs in the endoplasmic reticulum and the golgi apparatus. Proteins are synthesized by ribo ...
which typically involves covalently attaching certain groups to histone residues. Once added to the histone, these groups (directly or indirectly) elicit either a loose and open histone conformation, euchromatin
Euchromatin (also called "open chromatin") is a lightly packed form of chromatin ( DNA, RNA, and protein) that is enriched in genes, and is often (but not always) under active transcription. Euchromatin stands in contrast to heterochromatin, whi ...
, or a tight and closed histone conformation, heterochromatin
Heterochromatin is a tightly packed form of DNA or '' condensed DNA'', which comes in multiple varieties. These varieties lie on a continue between the two extremes of constitutive heterochromatin and facultative heterochromatin. Both play a rol ...
. Euchromatin marks active transcription and gene expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, protein or non-coding RNA, and ultimately affect a phenotype, as the final effect. T ...
, as the light packing of histones in this way allows entry for proteins involved in the transcription process. As such, the tightly packed heterochromatin marks the absence of current gene expression.
While there exist several distinct post-translational modifications for histone
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei. They act as spools around which DNA winds to create structural units called nucleosomes. Nucleosomes in turn a ...
s, the four most common histone modifications include acetylation
:
In organic chemistry, acetylation is an organic esterification reaction with acetic acid. It introduces an acetyl group into a chemical compound. Such compounds are termed ''acetate esters'' or simply '' acetates''. Deacetylation is the oppos ...
, methylation
In the chemical sciences, methylation denotes the addition of a methyl group on a substrate, or the substitution of an atom (or group) by a methyl group. Methylation is a form of alkylation, with a methyl group replacing a hydrogen atom. These ...
, phosphorylation
In chemistry, phosphorylation is the attachment of a phosphate group to a molecule or an ion. This process and its inverse, dephosphorylation, are common in biology and could be driven by natural selection. Text was copied from this source, wh ...
and ubiquitination
Ubiquitin is a small (8.6 kDa) regulatory protein found in most tissues of eukaryotic organisms, i.e., it is found ''ubiquitously''. It was discovered in 1975 by Gideon Goldstein and further characterized throughout the late 1970s and 1980s. Fo ...
. Histone-modifying enzymes that induce a modification (e.g., add a functional group
In organic chemistry, a functional group is a substituent or moiety in a molecule that causes the molecule's characteristic chemical reactions. The same functional group will undergo the same or similar chemical reactions regardless of the r ...
) are dubbed writers, while enzymes that revert modifications are dubbed erasers. Furthermore, there are many uncommon histone modifications including ''O''-GlcNAcylation, sumoylation
In molecular biology, SUMO (Small Ubiquitin-like Modifier) proteins are a family of small proteins that are covalently attached to and detached from other proteins in cells to modify their function. This process is called SUMOylation (sometimes w ...
, ADP-ribosylation
ADP-ribosylation is the addition of one or more ADP-ribose moieties to a protein. It is a reversible post-translational modification that is involved in many cellular processes, including cell signaling, DNA repair, gene regulation and apoptosis ...
, citrullination
Citrullination or deimination is the conversion of the amino acid arginine in a protein into the amino acid citrulline. Citrulline is not one of the 20 standard amino acids encoded by DNA in the genetic code. Instead, it is the result of a post-tr ...
and proline isomerization In epigenetics, proline isomerization is the effect that ''cis-trans'' isomerization of the amino acid proline has on the regulation of gene expression. Similar to aspartic acid, the amino acid proline has the rare property of being able to occupy ...
. For a detailed example of histone modifications in transcription regulation see RNA polymerase control by chromatin structure
RNA polymerase II (RNAP II and Pol II) is a multiprotein complex that transcribes DNA into precursors of messenger RNA (mRNA) and most small nuclear RNA (snRNA) and microRNA. It is one of the three RNAP enzymes found in the nucleus of eukaryotic ...
and table " Examples of histone modifications in transcriptional regulation".
Common histone modifications
The four common histone modifications and their respective writer and eraser enzymes are shown in the table below.Acetylation

Histone acetylation
Histone acetyltransferases (HATs) are enzymes that acetylate conserved lysine amino acids on histone proteins by transferring an acetyl group from acetyl-CoA to form ε-''N''-acetyllysine. DNA is wrapped around histones, and, by transferring an ...
, or the addition of an acetyl group
In organic chemistry, acetyl is a functional group with the chemical formula and the structure . It is sometimes represented by the symbol Ac (not to be confused with the element actinium). In IUPAC nomenclature, acetyl is called ethanoyl, ...
to histones, is facilitated by histone acetyltransferases (HATs) which target lysine (K) residues on the N-terminal
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the ami ...
histone tail. Histone deacetylases (HDACs) facilitate the removal of such groups. The positive charge on a histone is always neutralized upon acetylation, creating euchromatin
Euchromatin (also called "open chromatin") is a lightly packed form of chromatin ( DNA, RNA, and protein) that is enriched in genes, and is often (but not always) under active transcription. Euchromatin stands in contrast to heterochromatin, whi ...
which increases transcription and expression of the target gene. Lysine residues 9, 14, 18, and 23 of core histone H3 and residues 5, 8, 12, and 16 of H4 are all targeted for acetylation.
Methylation
Histone methylation
Histone methylation is a process by which methyl groups are transferred to amino acids of histone proteins that make up nucleosomes, which the DNA double helix wraps around to form chromosomes. Methylation of histones can either increase or decr ...
involves adding methyl group
In organic chemistry, a methyl group is an alkyl derived from methane, containing one carbon atom bonded to three hydrogen atoms, having chemical formula . In formulas, the group is often abbreviated as Me. This hydrocarbon group occurs in ma ...
s to histones, primarily on lysine (K) or arginine (R) residues. The addition and removal of methyl groups is carried out by histone methyltransferases (HMTs) and histone demethylase
Demethylases are enzymes that remove methyl (CH3) groups from nucleic acids, proteins (particularly histones), and other molecules. Demethylases are important epigenetics, epigenetic proteins, as they are responsible for transcriptional regulation ...
s (KDMs) respectively. Histone methylation is responsible for either activation or repression of genes, depending on the target site, and plays an important role in development and learning. Common histone methylations include H3K9 dimethylation and H3K27 trimethylation, which are both largely associated with gene silencing, whereas H3K4, H3K36, and H3K79 methylations are associated with an increase in gene transcription.
Phosphorylation
histone
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei. They act as spools around which DNA winds to create structural units called nucleosomes. Nucleosomes in turn a ...
. Protein kinases (PTKs) catalyze the phosphorylation of histones and protein phosphatases (PPs) catalyze the dephosphorylation of histones. Much like histone acetylation, histone phosphorylation neutralizes the negative charge on histones which induces euchromatin
Euchromatin (also called "open chromatin") is a lightly packed form of chromatin ( DNA, RNA, and protein) that is enriched in genes, and is often (but not always) under active transcription. Euchromatin stands in contrast to heterochromatin, whi ...
and increases gene expression. Histone phosphorylation occurs on serine (S), threonine (T) and tyrosine (Y) amino-acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although hundreds of amino acids exist in nature, by far the most important are the alpha-amino acids, which comprise proteins. Only 22 alpha ami ...
residues mainly in the N-terminal
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the ami ...
histone tails.
Additionally, the phosphorylation of histones has been found to play a role in DNA repair
DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as radiation can cause DNA d ...
and chromatin condensation
Prophase () is the first stage of cell division in both mitosis and meiosis. Beginning after interphase, DNA has already been replicated when the cell enters prophase. The main occurrences in prophase are the condensation of the chromatin reti ...
during cell division
Cell division is the process by which a parent cell divides into two daughter cells. Cell division usually occurs as part of a larger cell cycle in which the cell grows and replicates its chromosome(s) before dividing. In eukaryotes, there ...
. One such example is the phosphorylation of S139 on H2AX
H2A histone family member X (usually abbreviated as H2AX) is a type of histone protein from the H2A family encoded by the ''H2AFX'' gene. An important phosphorylated form is γH2AX (S139), which forms when double-strand breaks appear.
In humans ...
histones, which is needed to repair double-stranded break
DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as radiation can cause DNA dama ...
s in the DNA.
Ubiquitination
Ubiquitin
Ubiquitin is a small (8.6 kDa) regulatory protein found in most tissues of eukaryotic organisms, i.e., it is found ''ubiquitously''. It was discovered in 1975 by Gideon Goldstein and further characterized throughout the late 1970s and 1980s. Fo ...
ation is a post-translational modification involving the addition of ubiquitin proteins onto target proteins. Histones are often ubiquitinated with one ubiquitin molecule (monoubiquitination), but can also be modified with ubiquitin chains (polyubiquitination), both of which can have variable effects on gene transcription. Ubiquitin ligase
A ubiquitin ligase (also called an E3 ubiquitin ligase) is a protein that recruits an E2 ubiquitin-conjugating enzyme that has been loaded with ubiquitin, recognizes a protein substrate, and assists or directly catalyzes the transfer of ubiquit ...
s add these ubiquitin proteins while deubiquitinating enzymes (DUBs) remove these groups. Ubiquitination of the H2A core histone typically represses gene expression as it prevents methylation at H3K4, while H2B ubiquitination is necessary for H3K4 methylation and can lead to both gene activation or repression. Additionally, histone ubiquitination is related to genomic maintenance, as ubiquitination of histone H2AX is involved in DNA damage
DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as radiation can cause DNA d ...
recognition of DNA double-strand breaks.
Uncommon histone modifications
Additional infrequent histone modifications and their effects are listed in the table below.O-GlcNAcylation
 The presence of O-GlcNAcylation (O-GlcNAc) on serine (S) and threonine (T) histone residues is known to mediate other post-transcriptional histone modifications. The addition and removal of GlcNAc groups are performed by O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA) respectively. While our understanding of these processes is limited, GlcNAcylation of S112 on core histone H2B has been found to promote monoubiquitination of K120. Similarly, OGT associates with the HCF1 complex which interacts with
The presence of O-GlcNAcylation (O-GlcNAc) on serine (S) and threonine (T) histone residues is known to mediate other post-transcriptional histone modifications. The addition and removal of GlcNAc groups are performed by O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA) respectively. While our understanding of these processes is limited, GlcNAcylation of S112 on core histone H2B has been found to promote monoubiquitination of K120. Similarly, OGT associates with the HCF1 complex which interacts with BAP1
BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) is a deubiquitinating enzyme that in humans is encoded by the ''BAP1'' gene. ''BAP1'' encodes an 80.4 kDa nuclear-localizing protein with a ubiquitin carboxy-terminal hydrolase (U ...
to mediate deubiquitination of H2A. OGT is also involved in the trimethylation of H3K27 and creates a co-repressor complex to promote histone deacetylation upon binding to SIN3A
Paired amphipathic helix protein Sin3a is a protein that in humans is encoded by the ''SIN3A'' gene.
Function
The protein encoded by this gene is a transcriptional regulatory protein. It contains paired amphipathic helix (PAH) domains, which a ...
.
Sumoylation
SUMOylation
In molecular biology, SUMO (Small Ubiquitin-like Modifier) proteins are a family of small proteins that are covalently attached to and detached from other proteins in cells to modify their function. This process is called SUMOylation (sometimes w ...
refers to the addition of Small Ubiquitin-like Modifier (SUMO) proteins onto histones. SUMOylation involves covalent attachments between SUMO proteins and lysine (K) residues on histones and is carried out in three main steps by three respective enzymes: activation via SUMO E1, conjugation
Conjugation or conjugate may refer to:
Linguistics
*Grammatical conjugation, the modification of a verb from its basic form
* Emotive conjugation or Russell's conjugation, the use of loaded language
Mathematics
*Complex conjugation, the change ...
via SUMO E2, and ligation
Ligation may refer to:
* Ligation (molecular biology), the covalent linking of two ends of DNA or RNA molecules
* In medicine, the making of a ligature (tie)
* Chemical ligation, the production of peptides from amino acids
* Tubal ligation, a meth ...
via SUMO E3. In humans, SUMO E1 has been identified as the heterodimer
In biochemistry, a protein dimer is a macromolecular complex formed by two protein monomers, or single proteins, which are usually non-covalently bound. Many macromolecules, such as proteins or nucleic acids, form dimers. The word ''dimer'' ha ...
SAE1/ SAE2, SUMO E2 is known as UBE2I
SUMO-conjugating enzyme UBC9 is an enzyme that in humans is encoded by the ''UBE2I'' gene. It is also sometimes referred to as "ubiquitin conjugating enzyme E2I" or "ubiquitin carrier protein 9", even though these names do not accurately describ ...
, and the SUMO E3 role may be a multi-protein complex played by a handful of different enzymes.
SUMOylation affects the chromatin status (looseness) of the histone and influences the assembly of transcription factor
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The f ...
s on genetic promoters, leading to either transcriptional repression or activation depending on the substrate. SUMOylation also plays a role in the major DNA repair pathways of base excision repair
Base excision repair (BER) is a cellular mechanism, studied in the fields of biochemistry and genetics, that repairs damaged DNA throughout the cell cycle. It is responsible primarily for removing small, non-helix-distorting base lesions from ...
, nucleotide excision repair
Nucleotide excision repair is a DNA repair mechanism. DNA damage occurs constantly because of chemicals (e.g. intercalating agents), radiation and other mutagens. Three excision repair pathways exist to repair single stranded DNA damage: Nucle ...
, non-homologous end joining
Non-homologous end joining (NHEJ) is a pathway that repairs double-strand breaks in DNA. NHEJ is referred to as "non-homologous" because the break ends are directly ligated without the need for a homologous template, in contrast to homology direc ...
and homologous recombination
Homologous recombination is a type of genetic recombination in which genetic information is exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acids (usually DNA as in cellular organisms but may ...
repair. Additionally, SUMOylation facilitates error prone translesion synthesis
DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as radiation can cause DNA dama ...
.
ADP-ribosylation
protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, res ...
. ADPr is an important mechanism in gene regulation
Regulation of gene expression, or gene regulation, includes a wide range of mechanisms that are used by cells to increase or decrease the production of specific gene products ( protein or RNA). Sophisticated programs of gene expression are w ...
that affects chromatin organization, the binding of transcription factor
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The f ...
s, and mRNA processing through poly-ADP ribose polymerase (PARP) enzymes. There are multiple types of PARP proteins, but the subclass of DNA-dependent PARP proteins including PARP-1, PARP-2, and PARP-3 interact with the histone. The PARP-1 enzyme is the most prominent of these three proteins in the context of gene regulation and interacts with all five histone proteins.
Like PARPs 2 and 3, the catalytic activity of PARP-1 is activated by discontinuous DNA fragments, DNA fragments with single-stranded breaks. PARP-1 binds histones near the axis where DNA enters and exits the nucleosome and additionally interacts with numerous chromatin-associated proteins which allow for indirect association with chromatin. Upon binding to chromatin, PARP-1 produces repressive histone marks that can alter the conformational state of histones and inhibit gene expression so that DNA repair
DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as radiation can cause DNA d ...
can take place. Other avenues of transcription regulation by PARP-1 include acting as a transcription coregulator
In molecular biology and genetics, transcription coregulators are proteins that interact with transcription factors to either activate or repress the transcription of specific genes. Transcription coregulators that activate gene transcription ar ...
, regulation of RNA and modulation of DNA methylation
DNA methylation is a biological process by which methyl groups are added to the DNA molecule. Methylation can change the activity of a DNA segment without changing the sequence. When located in a gene promoter, DNA methylation typically acts ...
via inhibiting the DNA methyltransferase
In biochemistry, the DNA methyltransferase (DNA MTase, DNMT) family of enzymes catalyze the transfer of a methyl group to DNA. DNA methylation serves a wide variety of biological functions. All the known DNA methyltransferases use S-adenosyl m ...
Dnmt1
DNA (cytosine-5)-methyltransferase 1 is an enzyme that catalyzes the transfer of methyl groups to specific CpG structures in DNA, a process called DNA methylation. In humans, it is encoded by the ''DNMT1'' gene. DNMT1 forms part of the family of ...
.
Citrullination
Citrullination
Citrullination or deimination is the conversion of the amino acid arginine in a protein into the amino acid citrulline. Citrulline is not one of the 20 standard amino acids encoded by DNA in the genetic code. Instead, it is the result of a post-tr ...
, or deimination, is the process by which the amino acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although hundreds of amino acids exist in nature, by far the most important are the alpha-amino acids, which comprise proteins. Only 22 alpha ...
arginine (R) is converted into citrulline
The organic compound citrulline is an α-amino acid. Its name is derived from '' citrullus'', the Latin word for watermelon. Although named and described by gastroenterologists since the late 19th century, it was first isolated from watermelon in ...
. Protein arginine deiminases (PADs) replace the ketimine
In organic chemistry, an imine ( or ) is a functional group or organic compound containing a carbon– nitrogen double bond (). The nitrogen atom can be attached to a hydrogen or an organic group (R). The carbon atom has two additional si ...
group of arginine with a ketone
In organic chemistry, a ketone is a functional group with the structure R–C(=O)–R', where R and R' can be a variety of carbon-containing substituents. Ketones contain a carbonyl group –C(=O)– (which contains a carbon-oxygen double b ...
group to form the citrulline. PAD4 is the deaminase involved in histone modification and converts arginine to citrulline on histones H3 and H4; because arginine methylation
In the chemical sciences, methylation denotes the addition of a methyl group on a substrate, or the substitution of an atom (or group) by a methyl group. Methylation is a form of alkylation, with a methyl group replacing a hydrogen atom. These ...
on these histones is important for transcriptional activation, citrullination of certain residues can cause the eventual loss of methylation, leading to decreased gene transcription; specific citrullination of H3R2, H3R8, H3R17, and H3R26 residues have been identified in breast cancer cells. As of research conducted in 2019, this process is thought to be irreversible.
Proline Isomerization
Isomerization
In chemistry, isomerization or isomerisation is the process in which a molecule, polyatomic ion or molecular fragment is transformed into an isomer with a different chemical structure. Enolization is an example of isomerization, as is tautomeriz ...
involves transforming a molecule so that it adopts a different structural conformation; proline isomerization In epigenetics, proline isomerization is the effect that ''cis-trans'' isomerization of the amino acid proline has on the regulation of gene expression. Similar to aspartic acid, the amino acid proline has the rare property of being able to occupy ...
plays an integral role in the modification of histone tails. Fpr4 is the prolyl isomerase enzyme (PPIase) which converts the amino acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although hundreds of amino acids exist in nature, by far the most important are the alpha-amino acids, which comprise proteins. Only 22 alpha ...
proline (P) on histones between the cis and trans conformations. While Fpr4 has catalytic activity
Catalysis () is the process of increasing the rate of a chemical reaction by adding a substance known as a catalyst (). Catalysts are not consumed in the reaction and remain unchanged after it. If the reaction is rapid and the catalyst recyc ...
on a number of prolines on the N-terminal region of core histone H3 (P16, P30 and P38), it most readily binds to P38.
Near H3P38 lies near the lysine (K) residue H3K36, and changes in P38 can affect the methylation
In the chemical sciences, methylation denotes the addition of a methyl group on a substrate, or the substitution of an atom (or group) by a methyl group. Methylation is a form of alkylation, with a methyl group replacing a hydrogen atom. These ...
status of K36. The two possible P38 isomers available, cis and trans, cause differential effects that are opposite of each other. The cis position induces compact histones and decreases the ability of proteins to bind to the DNA, thus preventing methylation of K36 and decreasing gene transcription. Conversely, the trans position of P38 promotes a more open histone conformation, allowing for K36 methylation and leading to an increase gene transcription.
Role in research
Cancer
Alterations in the functions of histone-modifying enzymes deregulate the control of chromatin-based processes, ultimately leading tooncogenic
Carcinogenesis, also called oncogenesis or tumorigenesis, is the formation of a cancer, whereby normal cells are transformed into cancer cells. The process is characterized by changes at the cellular, genetic, and epigenetic levels and abnor ...
transformation and cancer
Cancer is a group of diseases involving abnormal cell growth with the potential to invade or spread to other parts of the body. These contrast with benign tumors, which do not spread. Possible signs and symptoms include a lump, abnormal b ...
. Both DNA methylation
DNA methylation is a biological process by which methyl groups are added to the DNA molecule. Methylation can change the activity of a DNA segment without changing the sequence. When located in a gene promoter, DNA methylation typically acts ...
and histone modifications show patterns of distribution in cancer cells. These epigenetic
In biology, epigenetics is the study of stable phenotypic changes (known as ''marks'') that do not involve alterations in the DNA sequence. The Greek prefix '' epi-'' ( "over, outside of, around") in ''epigenetics'' implies features that are ...
alterations may occur at different stages of tumourigenesis and thus contribute to both the development and/or progression of cancer.
Other Research
Vitamin B12
Vitamin B12, also known as cobalamin, is a water-soluble vitamin involved in metabolism. It is one of eight B vitamins. It is required by animals, which use it as a cofactor in DNA synthesis, in both fatty acid and amino acid metabolism. ...
deficiency in mice has been shown to alter expression of histone modifying enzymes in the brain, leading to behavioral changes and epigenetic
In biology, epigenetics is the study of stable phenotypic changes (known as ''marks'') that do not involve alterations in the DNA sequence. The Greek prefix '' epi-'' ( "over, outside of, around") in ''epigenetics'' implies features that are ...
reprogramming. Evidences also show the importance of HDACs in regulation of lipid metabolism
Lipid metabolism is the synthesis and degradation of lipids in cells, involving the breakdown or storage of fats for energy and the synthesis of structural and functional lipids, such as those involved in the construction of cell membranes. In anim ...
and other metabolic pathways playing a role in the pathophysiology
Pathophysiology ( physiopathology) – a convergence of pathology with physiology – is the study of the disordered physiological processes that cause, result from, or are otherwise associated with a disease or injury. Pathology is ...
of metabolic disorders.
See also
* DNA *Histone
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei. They act as spools around which DNA winds to create structural units called nucleosomes. Nucleosomes in turn a ...
*Nucleosome
A nucleosome is the basic structural unit of DNA packaging in eukaryotes. The structure of a nucleosome consists of a segment of DNA wound around eight histone proteins and resembles thread wrapped around a spool. The nucleosome is the fundame ...
*Chromatin
Chromatin is a complex of DNA and protein found in eukaryote, eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important ...
*Euchromatin
Euchromatin (also called "open chromatin") is a lightly packed form of chromatin ( DNA, RNA, and protein) that is enriched in genes, and is often (but not always) under active transcription. Euchromatin stands in contrast to heterochromatin, whi ...
*Heterochromatin
Heterochromatin is a tightly packed form of DNA or '' condensed DNA'', which comes in multiple varieties. These varieties lie on a continue between the two extremes of constitutive heterochromatin and facultative heterochromatin. Both play a rol ...
*Histone acetylation and deacetylation
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei. They act as spools around which DNA winds to create structural units called nucleosomes. Nucleosomes in turn are w ...
*Histone methylation
Histone methylation is a process by which methyl groups are transferred to amino acids of histone proteins that make up nucleosomes, which the DNA double helix wraps around to form chromosomes. Methylation of histones can either increase or decr ...
*Protein phosphorylation
Protein phosphorylation is a reversible post-translational modification of proteins in which an amino acid residue is phosphorylated by a protein kinase by the addition of a covalently bound phosphate group. Phosphorylation alters the structura ...
*Ubiquitin
Ubiquitin is a small (8.6 kDa) regulatory protein found in most tissues of eukaryotic organisms, i.e., it is found ''ubiquitously''. It was discovered in 1975 by Gideon Goldstein and further characterized throughout the late 1970s and 1980s. Fo ...
*O-GlcNAc
''O''-GlcNAc (short for ''O''-linked GlcNAc or ''O''-linked β-''N''-acetylglucosamine) is a reversible enzymatic post-translational modification that is found on serine and threonine residues of nucleocytoplasmic proteins. The modification is c ...
*Sumoylation
In molecular biology, SUMO (Small Ubiquitin-like Modifier) proteins are a family of small proteins that are covalently attached to and detached from other proteins in cells to modify their function. This process is called SUMOylation (sometimes w ...
*ADP-ribosylation
ADP-ribosylation is the addition of one or more ADP-ribose moieties to a protein. It is a reversible post-translational modification that is involved in many cellular processes, including cell signaling, DNA repair, gene regulation and apoptosis ...
*Citrullination
Citrullination or deimination is the conversion of the amino acid arginine in a protein into the amino acid citrulline. Citrulline is not one of the 20 standard amino acids encoded by DNA in the genetic code. Instead, it is the result of a post-tr ...
* Proline isomerization in epigenetics
References
{{DEFAULTSORT:Histone-Modifying Enzymes Epigenetics Proteins