frame shift on:
[Wikipedia]
[Google]
[Amazon]
Ribosomal frameshifting, also known as translational frameshifting or translational recoding, is a biological phenomenon that occurs during
 In −1 frameshifting, the ribosome slips back one nucleotide and continues translation in the −1 frame. There are typically three elements that comprise a −1 frameshift signal: a
In −1 frameshifting, the ribosome slips back one nucleotide and continues translation in the −1 frame. There are typically three elements that comprise a −1 frameshift signal: a
 The slippery sequence for a +1 frameshift signal does not have the same motif, and instead appears to function by pausing the ribosome at a sequence encoding a rare amino acid. Ribosomes do not translate proteins at a steady rate, regardless of the sequence. Certain codons take longer to translate, because there are not equal amounts of
The slippery sequence for a +1 frameshift signal does not have the same motif, and instead appears to function by pausing the ribosome at a sequence encoding a rare amino acid. Ribosomes do not translate proteins at a steady rate, regardless of the sequence. Certain codons take longer to translate, because there are not equal amounts of
Wise2
— aligns a
FastY
— compare a DNA sequence to a
Path
— tool that compares two
Recode2
— Database of recoded genes, including those that require programmed Translational frameshift. {{Use dmy dates, date=September 2019 RNA Gene expression Cis-regulatory RNA elements Genetics
translation
Translation is the communication of the meaning of a source-language text by means of an equivalent target-language text. The English language draws a terminological distinction (which does not exist in every language) between ''transla ...
that results in the production of multiple, unique protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, res ...
s from a single mRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
mRNA is created during the ...
. The process can be programmed by the nucleotide sequence of the mRNA and is sometimes affected by the secondary, 3-dimensional mRNA structure. It has been described mainly in viruses
A virus is a submicroscopic infectious agent that replicates only inside the living cells of an organism. Viruses infect all life forms, from animals and plants to microorganisms, including bacteria and archaea.
Since Dmitri Ivanovsky's ...
(especially retroviruses), retrotransposons
Retrotransposons (also called Class I transposable elements or transposons via RNA intermediates) are a type of genetic component that copy and paste themselves into different genomic locations ( transposon) by converting RNA back into DNA throug ...
and bacterial insertion elements, and also in some cellular genes.
Process overview
Proteins are translated by reading tri-nucleotides on the mRNA strand, also known as codons, from one end of themRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
mRNA is created during the ...
to the other (from the 5' to the 3' end) starting with the amino acid methionine as the start (initiation) codon AUG. Each codon is translated into a single amino acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although hundreds of amino acids exist in nature, by far the most important are the alpha-amino acids, which comprise proteins. Only 22 alpha a ...
. The code itself is considered degenerate, meaning that a particular amino acid can be specified by more than one codons. However, a shift of any number of nucleotides that is not divisible by 3 in the reading frame will result in subsequent codons to be read differently. This effectively changes the ribosomal reading frame
In molecular biology, a reading frame is a way of dividing the sequence of nucleotides in a nucleic acid ( DNA or RNA) molecule into a set of consecutive, non-overlapping triplets. Where these triplets equate to amino acids or stop signals during ...
.
Sentence example
In this example, the following sentence with three-letter words makes sense when read from the beginning: , Start, THE CAT AND THE MAN ARE FAT ... , Start, 123 123 123 123 123 123 123 ... However, if the reading frame is shifted by one letter to between the T and H of the first word (effectively a +1 frameshift when considering the 0 position to be the initial position of T), T, Start, HEC ATA NDT HEM ANA REF AT... -, Start, 123 123 123 123 123 123 12... then the sentence reads differently, making no sense.DNA example
In this example, the following sequence is a region of the human mitochondrial genome with the two overlapping genesMT-ATP8
''MT-ATP8'' (or ''ATP8'') is a mitochondrial gene with the full name 'mitochondrially encoded ATP synthase membrane subunit 8' that encodes a subunit of mitochondrial ATP synthase, ATP synthase Fo subunit 8 (or subunit A6L). This subunit be ...
and MT-ATP6
''MT-ATP6'' (or ''ATP6'') is a mitochondrial gene with the full name 'mitochondrially encoded ATP synthase membrane subunit 6' that encodes the ATP synthase Fo subunit 6 (or subunit/chain A). This subunit belongs to the Fo complex of the large, ...
.
When read from the beginning, these codons make sense to a ribosome and can be translated into amino acids (AA) under the vertebrate mitochondrial code:
, Start, AAC GAA AAT CTG TTC GCT TCA ...
, Start, 123 123 123 123 123 123 123 ...
, AA , N E N L F A S ...
However, let's change the reading frame by starting one nucleotide downstream (effectively a "+1 frameshift" when considering the 0 position to be the initial position of A):
A, Start, ACG AAA ATC TGT TCG CTT CA...
-, Start, 123 123 123 123 123 123 12...
, AA , T K I C S L ...
Now, because of this +1 frameshifting, the DNA sequence is read differently. The different codon reading frame therefore yields different amino acids.
In the case of a translating ribosome, a frameshift can either result in nonsense
Nonsense is a communication, via speech, writing, or any other symbolic system, that lacks any coherent meaning. Sometimes in ordinary usage, nonsense is synonymous with absurdity or the ridiculous. Many poets, novelists and songwriters have u ...
(a premature stop codon) after the frameshift, or the creation of a completely new protein after the frameshift. In the case where a frameshift results in nonsense, the NMD (nonsense-mediated mRNA decay
Nonsense-mediated mRNA decay (NMD) is a surveillance pathway that exists in all eukaryotes. Its main function is to reduce errors in gene expression by eliminating mRNA transcripts that contain premature stop codons. Translation of these aberran ...
) pathway may destroy the mRNA transcript, so frameshifting would serve as a method of regulating the expression level of the associated gene.
Function
In viruses this phenomenon may be programmed to occur at particular sites and allows the virus to encode multiple types of proteins from the same mRNA. Notable examples includeHIV-1
The subtypes of HIV include two major types, HIV type 1 (HIV-1) and HIV type 2 (HIV-2). HIV-1 is related to viruses found in chimpanzees and gorillas living in western Africa, while HIV-2 viruses are related to viruses found in the sooty mangabey ...
(human immunodeficiency virus), RSV ( Rous sarcoma virus) and the influenza virus (flu), which all rely on frameshifting to create a proper ratio of 0-frame (normal translation) and "trans-frame" (encoded by frameshifted sequence) proteins. Its use in viruses is primarily for compacting more gene
In biology, the word gene (from , ; "... Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a b ...
tic information into a shorter amount of genetic material.
In eukaryotes it appears to play a role in regulating gene expression levels by generating premature stops and producing nonfunctional transcripts.
Types of frameshifting
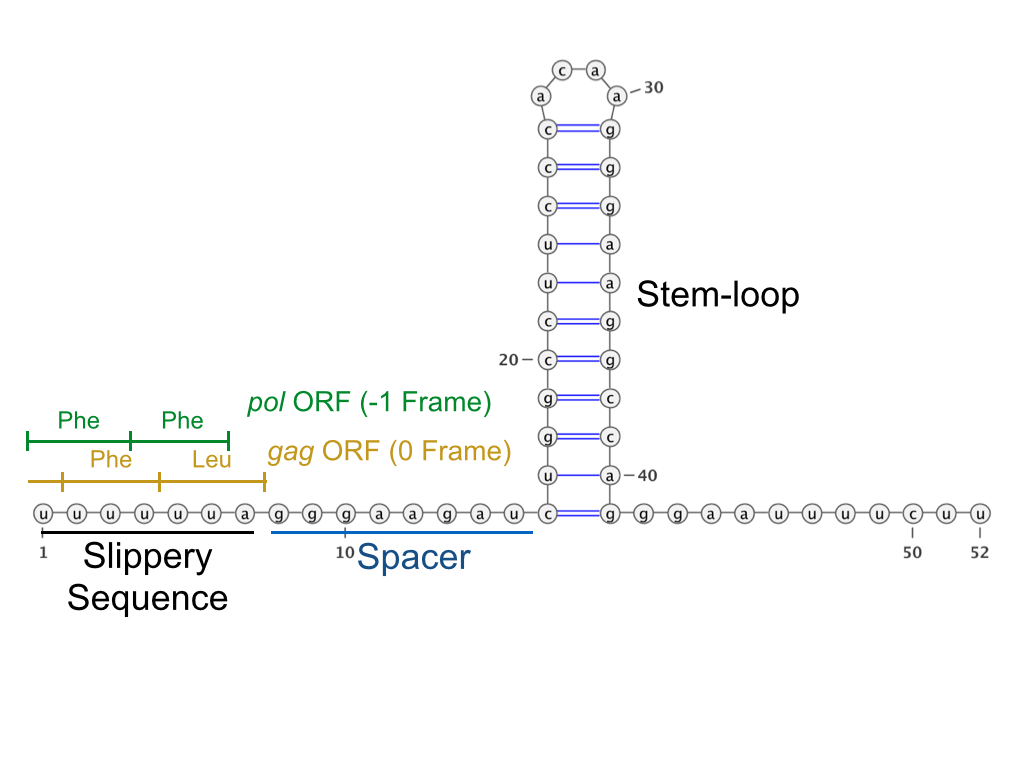
The most common type of frameshifting is −1 frameshifting or programmed −1 ribosomal frameshifting (−1 PRF). Other, rarer types of frameshifting include +1 and −2 frameshifting. −1 and +1 frameshifting are believed to be controlled by different mechanisms, which are discussed below. Both mechanisms are kinetically driven.Programmed −1 ribosomal frameshifting
 In −1 frameshifting, the ribosome slips back one nucleotide and continues translation in the −1 frame. There are typically three elements that comprise a −1 frameshift signal: a
In −1 frameshifting, the ribosome slips back one nucleotide and continues translation in the −1 frame. There are typically three elements that comprise a −1 frameshift signal: a slippery sequence
A slippery sequence is a small section of codon nucleotide sequences (usually UUUAAAC) that controls the rate and chance of ribosomal frameshifting. A slippery sequence causes a faster ribosomal transfer which in turn can cause the reading ribosom ...
, a spacer region, and an RNA secondary structure. The slippery sequence fits a X_XXY_YYH motif, where XXX is any three identical nucleotides (though some exceptions occur), YYY typically represents UUU or AAA, and H is A, C or U. Because the structure of this motif contains 2 adjacent 3-nucleotide repeats it is believed that −1 frameshifting is described by a tandem slippage model, in which the ribosomal P-site tRNA anticodon re-pairs from XXY to XXX and the A-site anticodon re-pairs from YYH to YYY simultaneously. These new pairings are identical to the 0-frame pairings except at their third positions. This difference does not significantly disfavor anticodon binding because the third nucleotide in a codon, known as the wobble position
A wobble base pair is a pairing between two nucleotides in RNA molecules that does not follow Watson-Crick base pair rules. The four main wobble base pairs are guanine-uracil (G-U), hypoxanthine-uracil (I-U), hypoxanthine-adenine (I-A), and hyp ...
, has weaker tRNA anticodon binding specificity than the first and second nucleotides. In this model, the motif structure is explained by the fact that the first and second positions of the anticodons must be able to pair perfectly in both the 0 and −1 frames. Therefore, nucleotides 2 and 1 must be identical, and nucleotides 3 and 2 must also be identical, leading to a required sequence of 3 identical nucleotides for each tRNA that slips.
+1 ribosomal frameshifting
 The slippery sequence for a +1 frameshift signal does not have the same motif, and instead appears to function by pausing the ribosome at a sequence encoding a rare amino acid. Ribosomes do not translate proteins at a steady rate, regardless of the sequence. Certain codons take longer to translate, because there are not equal amounts of
The slippery sequence for a +1 frameshift signal does not have the same motif, and instead appears to function by pausing the ribosome at a sequence encoding a rare amino acid. Ribosomes do not translate proteins at a steady rate, regardless of the sequence. Certain codons take longer to translate, because there are not equal amounts of tRNA
Transfer RNA (abbreviated tRNA and formerly referred to as sRNA, for soluble RNA) is an adaptor molecule composed of RNA, typically 76 to 90 nucleotides in length (in eukaryotes), that serves as the physical link between the mRNA and the amino ...
of that particular codon in the cytosol
The cytosol, also known as cytoplasmic matrix or groundplasm, is one of the liquids found inside cells ( intracellular fluid (ICF)). It is separated into compartments by membranes. For example, the mitochondrial matrix separates the mitochondri ...
. Due to this lag, there exist in small sections of codons sequences that control the rate of ribosomal frameshifting. Specifically, the ribosome must pause to wait for the arrival of a rare tRNA, and this increases the kinetic favorability of the ribosome and its associated tRNA slipping into the new frame. In this model, the change in reading frame is caused by a single tRNA slip rather than two.
Controlling mechanisms
Ribosomal frameshifting may be controlled by mechanisms found in the mRNA sequence (cis-acting). This generally refers to a slippery sequence, a RNA secondary structure, or both. A −1 frameshift signal consists of both elements separated by a spacer region typically 5–9 nucleotides long. Frameshifting may also be induced by other molecules which interact with the ribosome or the mRNA (trans-acting).Frameshift signal elements

Slippery sequence
Slippery sequence
A slippery sequence is a small section of codon nucleotide sequences (usually UUUAAAC) that controls the rate and chance of ribosomal frameshifting. A slippery sequence causes a faster ribosomal transfer which in turn can cause the reading ribosom ...
s can potentially make the reading ribosome "slip" and skip a number of nucleotides
Nucleotides are organic molecules consisting of a nucleoside and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of which are essential biomolecules with ...
(usually only 1) and read a completely different frame thereafter. In programmed −1 ribosomal frameshifting, the slippery sequence fits a X_XXY_YYH motif, where XXX is any three identical nucleotides (though some exceptions occur), YYY typically represents UUU or AAA, and H is A, C or U. In the case of +1 frameshifting, the slippery sequence contains codons for which the corresponding tRNA is more rare, and the frameshift is favored because the codon in the new frame has a more common associated tRNA. One example of a slippery sequence is the polyA
Polyadenylation is the addition of a poly(A) tail to an RNA transcript, typically a messenger RNA (mRNA). The poly(A) tail consists of multiple adenosine monophosphates; in other words, it is a stretch of RNA that has only adenine bases. In euka ...
on mRNA, which is known to induce ribosome slippage even in the absence of any other elements.
RNA secondary structure
Efficient ribosomal frameshifting generally requires the presence of an RNA secondary structure to enhance the effects of the slippery sequence. The RNA structure (which can be a stem-loop or pseudoknot) is thought to pause the ribosome on the slippery site during translation, forcing it to relocate and continue replication from the −1 position. It is believed that this occurs because the structure physically blocks movement of the ribosome by becoming stuck in the ribosome mRNA tunnel. This model is supported by the fact that strength of the pseudoknot has been positively correlated with the level of frameshifting for associated mRNA. Below are examples of predicted secondary structures for frameshift elements shown to stimulate frameshifting in a variety of organisms. The majority of the structures shown are stem-loops, with the exception of the ALIL (apical loop-internal loop) pseudoknot structure. In these images, the larger and incomplete circles of mRNA represent linear regions. The secondary "stem-loop" structures, where "stems" are formed by a region of mRNA base pairing with another region on the same strand, are shown protruding from the linear DNA. The linear region of the HIV ribosomal frameshift signal contains a highly conserved UUU UUU A slippery sequence; many of the other predicted structures contain candidates for slippery sequences as well. The mRNA sequences in the images can be read according to a set of guidelines. While A, T, C, and G represent a particular nucleotide at a position, there are also letters that represent ambiguity which are used when more than one kind of nucleotide could occur at that position. The rules of the International Union of Pure and Applied Chemistry (IUPAC
The International Union of Pure and Applied Chemistry (IUPAC ) is an international federation of National Adhering Organizations working for the advancement of the chemical sciences, especially by developing nomenclature and terminology. It is ...
) are as follows:
These symbols are also valid for RNA, except with U (uracil) replacing T (thymine).
Trans-acting elements
Small molecules, proteins, and nucleic acids have been found to stimulate levels of frameshifting. For example, the mechanism of a negative feedback loop in the polyamine synthesis pathway is based on polyamine levels stimulating an increase in +1 frameshifts, which results in production of an inhibitory enzyme. Certain proteins which are needed for codon recognition or which bind directly to the mRNA sequence have also been shown to modulate frameshifting levels.MicroRNA
MicroRNA (miRNA) are small, single-stranded, non-coding RNA molecules containing 21 to 23 nucleotides. Found in plants, animals and some viruses, miRNAs are involved in RNA silencing and post-transcriptional regulation of gene expression. mi ...
(miRNA) molecules may hybridize to a RNA secondary structure and affect its strength.
See also
* Antizyme RNA frameshifting stimulation element * Coronavirus frameshifting stimulation element * DnaX ribosomal frameshifting element * Frameshift mutation *HIV ribosomal frameshift signal
HIV ribosomal frameshift signal is a ribosomal frameshift (PRF) that human immunodeficiency virus (HIV) uses to translate several different proteins from the same sequence.
Intact and consistent protein biosynthesis relies on the ability of the ...
* Insertion sequence IS1222 ribosomal frameshifting element
* Recode database
* Ribosomal pause
* Slippery sequence
A slippery sequence is a small section of codon nucleotide sequences (usually UUUAAAC) that controls the rate and chance of ribosomal frameshifting. A slippery sequence causes a faster ribosomal transfer which in turn can cause the reading ribosom ...
References
External links
*Wise2
— aligns a
protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, res ...
against a DNA sequence allowing frameshift
Ribosomal frameshifting, also known as translational frameshifting or translational recoding, is a biological phenomenon that occurs during translation that results in the production of multiple, unique proteins from a single mRNA. The process can ...
s and introns
FastY
— compare a DNA sequence to a
protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, res ...
sequence database, allowing gaps and frameshift
Ribosomal frameshifting, also known as translational frameshifting or translational recoding, is a biological phenomenon that occurs during translation that results in the production of multiple, unique proteins from a single mRNA. The process can ...
s
Path
— tool that compares two
frameshift
Ribosomal frameshifting, also known as translational frameshifting or translational recoding, is a biological phenomenon that occurs during translation that results in the production of multiple, unique proteins from a single mRNA. The process can ...
proteins (back-translation
Translation is the communication of the meaning of a source-language text by means of an equivalent target-language text. The English language draws a terminological distinction (which does not exist in every language) between ''transla ...
principle)
Recode2
— Database of recoded genes, including those that require programmed Translational frameshift. {{Use dmy dates, date=September 2019 RNA Gene expression Cis-regulatory RNA elements Genetics