endonuclease on:
[Wikipedia]
[Google]
[Amazon]
Endonucleases are enzymes that cleave the
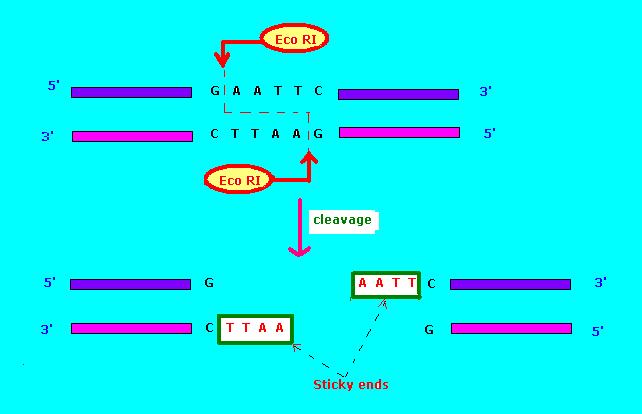 Restriction endonucleases come in several types. A restriction endonuclease typically requires a recognition site and a cleavage pattern (typically of nucleotide bases: A, C, G, T). If the recognition site is outside the region of the cleavage pattern, then the restriction endonuclease is referred to as Type I. If the recognition sequence overlaps with the cleavage sequence, then the restriction endonuclease
Restriction endonucleases come in several types. A restriction endonuclease typically requires a recognition site and a cleavage pattern (typically of nucleotide bases: A, C, G, T). If the recognition site is outside the region of the cleavage pattern, then the restriction endonuclease is referred to as Type I. If the recognition sequence overlaps with the cleavage sequence, then the restriction endonuclease

phosphodiester bond
In chemistry, a phosphodiester bond occurs when exactly two of the hydroxyl groups () in phosphoric acid react with hydroxyl groups on other molecules to form two ester bonds. The "bond" involves this linkage . Discussion of phosphodiesters is ...
within a polynucleotide chain. Some, such as deoxyribonuclease I, cut DNA relatively nonspecifically (without regard to sequence), while many, typically called restriction endonucleases
A restriction enzyme, restriction endonuclease, REase, ENase or'' restrictase '' is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class ...
or restriction enzymes, cleave only at very specific nucleotide sequences. Endonucleases differ from exonuclease
Exonucleases are enzymes that work by cleaving nucleotides one at a time from the end (exo) of a polynucleotide chain. A hydrolyzing reaction that breaks phosphodiester bonds at either the 3′ or the 5′ end occurs. Its close relative is ...
s, which cleave the ends of recognition sequences instead of the middle (endo) portion. Some enzymes known as "exo-endonucleases", however, are not limited to either nuclease function, displaying qualities that are both endo- and exo-like. Evidence suggests that endonuclease activity experiences a lag compared to exonuclease activity.
Restriction enzyme
A restriction enzyme, restriction endonuclease, REase, ENase or'' restrictase '' is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class ...
s are endonucleases from eubacteria
Bacteria (; singular: bacterium) are ubiquitous, mostly free-living organisms often consisting of one biological cell. They constitute a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria were amon ...
and archaea that recognize a specific DNA sequence. The nucleotide sequence recognized for cleavage by a restriction enzyme is called the restriction site. Typically, a restriction site will be a palindromic sequence about four to six nucleotides long. Most restriction endonucleases cleave the DNA strand unevenly, leaving complementary single-stranded ends. These ends can reconnect through hybridization and are termed "sticky ends". Once paired, the phosphodiester bonds of the fragments can be joined by DNA ligase
DNA ligase is a specific type of enzyme, a ligase, () that facilitates the joining of DNA strands together by catalyzing the formation of a phosphodiester bond. It plays a role in repairing single-strand breaks in duplex DNA in living orga ...
. There are hundreds of restriction endonucleases known, each attacking a different restriction site. The DNA fragments cleaved by the same endonuclease can be joined regardless of the origin of the DNA. Such DNA is called recombinant DNA
Recombinant DNA (rDNA) molecules are DNA molecules formed by laboratory methods of genetic recombination (such as molecular cloning) that bring together genetic material from multiple sources, creating sequences that would not otherwise be f ...
; DNA formed by the joining of genes into new combinations. ''Restriction endonucleases'' (restriction enzyme
A restriction enzyme, restriction endonuclease, REase, ENase or'' restrictase '' is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class ...
s) are divided into three categories, Type I, Type II, and Type III, according to their mechanism of action. These enzymes are often used in genetic engineering
Genetic engineering, also called genetic modification or genetic manipulation, is the modification and manipulation of an organism's genes using technology. It is a set of technologies used to change the genetic makeup of cells, including ...
to make recombinant DNA
Recombinant DNA (rDNA) molecules are DNA molecules formed by laboratory methods of genetic recombination (such as molecular cloning) that bring together genetic material from multiple sources, creating sequences that would not otherwise be f ...
for introduction into bacterial, plant, or animal cells, as well as in synthetic biology. One of the more famous endonucleases is Cas9
Cas9 (CRISPR associated protein 9, formerly called Cas5, Csn1, or Csx12) is a 160 kilodalton protein which plays a vital role in the immunological defense of certain bacteria against DNA viruses and plasmids, and is heavily utilized in gene ...
.
Categories
Ultimately, there are three categories ofrestriction endonucleases
A restriction enzyme, restriction endonuclease, REase, ENase or'' restrictase '' is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class ...
that relatively contribute to the cleavage of specific sequences. The types I and III are large multisubunit complexes that include both the endonucleases
Endonucleases are enzymes that cleave the phosphodiester bond within a polynucleotide chain. Some, such as deoxyribonuclease I, cut DNA relatively nonspecifically (without regard to sequence), while many, typically called restriction endonucleases ...
and methylase activities. Type I can cleave at random sites of about 1000 base pairs or more from the recognition sequence and it requires ATP as source of energy. Type II behaves slightly differently and was first isolated by Hamilton Smith in 1970. They are simpler versions of the endonucleases and require no ATP in their degradation processes. Some examples of type II restriction endonucleases include ''Bam''HI, ''Eco''RI, ''Eco''RV, ''Hin''dIII, and ''Hae''III. Type III, however, cleaves the DNA at about 25 base pairs from the recognition sequence and also requires ATP in the process.
Notations
The commonly used notation for restriction endonucleases is of the form "''Vwx''yZ", where "''Vwx''" are, in italics, the first letter of the genus and the first two letters of the species where this restriction endonuclease may be found, for example, ''Escherichia coli'', ''Eco'', and ''Haemophilus influenzae'', ''Hin''. This is followed by the optional, non-italicized symbol "y", which indicates the type or strain identification, for example, ''Eco''R for ''E. coli'' strains bearing the drug resistance transfer factor RTF-1, ''Eco''B for ''E. coli'' strain B, and ''Hin''d for ''H. influenzae'' strain d. Finally, when a particular type or strain has several different restriction endonucleases, these are identified by Roman numerals, thus, the restriction endonucleases from ''H. influenzae'' strain d are named ''Hin''dI, ''Hin''dII, ''Hin''dIII, etc. Another example: "''Hae''II" and "''Hae''III" refer to bacterium ''Haemophilus aegyptius'' (strain not specified), restriction endonucleases number II and number III, respectively. The restriction enzymes used in molecular biology usually recognize short target sequences of about 4 – 8 base pairs. For instance, the ''Eco''RI enzyme recognizes and cleaves the sequence 5' – GAATTC – 3'.restriction enzyme
A restriction enzyme, restriction endonuclease, REase, ENase or'' restrictase '' is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class ...
is Type II.
Further discussion
Restriction endonucleases may be found that cleave standard dsDNA (double-stranded DNA), or ssDNA (single-stranded DNA), or even RNA. This discussion is restricted to dsDNA; however, the discussion can be extended to the following: * Standard dsDNA * Non-standard DNA # Holliday junctions # Triple-stranded DNA, quadruple-stranded DNA (G-quadruplex
In molecular biology, G-quadruplex secondary structures (G4) are formed in nucleic acids by sequences that are rich in guanine. They are helical in shape and contain guanine tetrads that can form from one, two or four strands. The unimolecular ...
)
# Double-stranded hybrids of DNA and RNA (one strand is DNA, the other strand is RNA)
# Synthetic or artificial DNA (for example, containing bases other than A, C, G, T, refer to the work of Eric T. Kool). Research with synthetic codons, refer to the research by S. Benner, and enlarging the amino acid set in polypeptides, thus enlarging the proteome or proteomics, see the research by P. Schultz.
In addition, research is now underway to construct synthetic or artificial restriction endonucleases, especially with recognition sites that are unique within a genome.
Restriction endonucleases or restriction enzymes
A restriction enzyme, restriction endonuclease, REase, ENase or'' restrictase '' is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class ...
typically cleave in two ways: blunt-ended or sticky-ended patterns. An example of a Type I restriction endonuclease.
Furthermore, there exist DNA/RNA non-specific endonuclease
In molecular biology, enzymes in the DNA/RNA non-specific endonuclease family of bacterial and eukaryotic endonucleases share the following characteristics: they act on both DNA and RNA, cleave double-stranded and single-stranded nucleic aci ...
s, such as those that are found in ''Serratia marcescens
''Serratia marcescens'' () is a species of rod-shaped, Gram-negative bacteria in the family Yersiniaceae. It is a facultative anaerobe and an opportunistic pathogen in humans. It was discovered in 1819 by Bartolomeo Bizio in Padua, Italy.Serr ...
'', which act on dsDNA, ssDNA, and RNA.
DNA repair
Endonucleases play a role in DNA repair.AP endonuclease
Apurinic/apyrimidinic (AP) endonuclease is an enzyme that is involved in the DNA base excision repair pathway (BER). Its main role in the repair of damaged or mismatched nucleotides in DNA is to create a nick in the phosphodiester backbone of t ...
, specifically, catalyzes the incision of DNA exclusively at AP sites, and therefore prepares DNA for subsequent excision, repair synthesis and DNA ligation. For example, when depurination occurs, this lesion leaves a deoxyribose sugar with a missing base. The AP endonuclease recognizes this sugar and essentially cuts the DNA at this site and then allows for DNA repair to continue. ''E. coli'' cells contain two AP endonucleases: endonuclease IV (endoIV) and exonuclease III (exoIII) while in eukaryotes, there is only one AP endonuclease.

DNA crosslink repair
Repair of DNA in which the two complementary strands are joined by an interstrand covalent crosslink requires multiple incisions in order to disengage the strands and remove the damage. Incisions are required on both sides of the crosslink and on both strands of the duplex DNA. In mouse embryonic stem cells, an intermediate stage of crosslink repair involves production of double-strand breaks.Hanada K, Budzowska M, Modesti M, Maas A, Wyman C, Essers J, Kanaar R. The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. EMBO J. 2006 Oct 18;25(20):4921-32. doi: 10.1038/sj.emboj.7601344. Epub 2006 Oct 12. PMID: 17036055; PMCID: PMC1618088 MUS81/ EME1 is a structure specific endonuclease involved in converting interstrand crosslinks to double-strand breaks in a DNA replication-dependent manner. After introduction of a double-strand break, further steps are required to complete the repair process. If a crosslink is not properly repaired it can blockDNA replication
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all living organisms acting as the most essential part for biological inheritan ...
.
Thymine dimer repair
Exposure of bacteriophage (phage) T4 toultraviolet
Ultraviolet (UV) is a form of electromagnetic radiation with wavelength from 10 nm (with a corresponding frequency around 30 PHz) to 400 nm (750 THz), shorter than that of visible light, but longer than X-rays. UV radiation ...
irradiation induces thymine dimers in the phage DNA. The phage T4 ''denV'' gene encodes endonuclease V that catalyzes the initial steps in the repair of these UV-induced thymine dimers. Endonuclease V first cleaves the glycosylic bond on the 5’ side of a pyrimidine dimer and then catalyzes cleavage of the DNA phospodiester bond that originally linked the two nucleotides of the dimer. Subsequent steps in the repair process involve removal of the dimer remnants and repair synthesis to fill in the resulting single-strand gap using the undamaged strand as template.
Common endonucleases
Below are tables of common prokaryotic and eukaryotic endonucleases.Mutations
Xeroderma pigmentosa is a rare, autosomal recessive disease caused by a defective UV-specific endonuclease. Patients with mutations are unable to repair DNA damage caused by sunlight. Sickle Cell anemia is a disease caused by a point mutation. The sequence altered by the mutation eliminates the recognition site for the restriction endonuclease MstII that recognizes the nucleotide sequence. tRNA splicing endonuclease mutations cause pontocerebellar hypoplasia. Pontocerebellar hypoplasias (PCH) represent a group of neurodegenerative autosomal recessive disorders that is caused by mutations in three of the four different subunits of the tRNA-splicing endonuclease complex.See also
*Exonuclease
Exonucleases are enzymes that work by cleaving nucleotides one at a time from the end (exo) of a polynucleotide chain. A hydrolyzing reaction that breaks phosphodiester bonds at either the 3′ or the 5′ end occurs. Its close relative is ...
* Restriction endonuclease
* Nuclease
* Ribonuclease
Ribonuclease (commonly abbreviated RNase) is a type of nuclease that catalyzes the degradation of RNA into smaller components. Ribonucleases can be divided into endoribonucleases and exoribonucleases, and comprise several sub-classes within the ...
* AP endonuclease
Apurinic/apyrimidinic (AP) endonuclease is an enzyme that is involved in the DNA base excision repair pathway (BER). Its main role in the repair of damaged or mismatched nucleotides in DNA is to create a nick in the phosphodiester backbone of t ...
* HUH-tag
HUH endonucleases (HUH-tags) are sequence-specific single-stranded DNA (ssDNA) binding proteins originating from numerous species of bacteria and viruses. Viral HUH endonucleases are involved in initiating rolling circle replication while ones of ...
References
{{Portal bar, Biology, border=no EC 3.1