 Tet Repressor proteins (otherwise known as TetR) are proteins playing an important role in conferring
Tet Repressor proteins (otherwise known as TetR) are proteins playing an important role in conferring antibiotic resistance
Antimicrobial resistance (AMR) occurs when microbes evolve mechanisms that protect them from the effects of antimicrobials. All classes of microbes can evolve resistance. Fungi evolve antifungal resistance. Viruses evolve antiviral resistanc ...
to large categories of bacterial species.
Tetracycline
Tetracycline, sold under various brand names, is an oral antibiotic in the tetracyclines family of medications, used to treat a number of infections, including acne, cholera, brucellosis, plague, malaria, and syphilis.
Common side effects ...
(Tc) is a broad family of antibiotics to which bacteria have evolved resistance. Tc normally kills bacteria by binding to the bacterial ribosome and halting protein synthesis. The expression of Tc resistance genes is regulated by the repressor
In molecular genetics, a repressor is a DNA- or RNA-binding protein that inhibits the expression of one or more genes by binding to the operator or associated silencers. A DNA-binding repressor blocks the attachment of RNA polymerase to the ...
TetR. TetR represses the expression of TetA, a membrane protein that pumps out substances toxic to the bacteria like Tc, by binding the ''tetA'' operator
Operator may refer to:
Mathematics
* A symbol indicating a mathematical operation
* Logical operator or logical connective in mathematical logic
* Operator (mathematics), mapping that acts on elements of a space to produce elements of another ...
. In Tc-resistant bacteria, TetA will pump out Tc before it can bind to the ribosome because the repressive action of TetR on TetA is halted by binding of Tc to TetR. Therefore, TetR may have an important role in helping scientists to better understand mechanisms of antibiotic resistance
Antimicrobial resistance (AMR) occurs when microbes evolve mechanisms that protect them from the effects of antimicrobials. All classes of microbes can evolve resistance. Fungi evolve antifungal resistance. Viruses evolve antiviral resistanc ...
and how to treat antibiotic resistant bacteria. TetR is one of many proteins in the TetR protein family
A protein family is a group of evolutionarily related proteins. In many cases, a protein family has a corresponding gene family, in which each gene encodes a corresponding protein with a 1:1 relationship. The term "protein family" should not be c ...
, which is so named because TetR is the most well characterized member.
TetR is used in artificially engineered gene regulatory network
A gene (or genetic) regulatory network (GRN) is a collection of molecular regulators that interact with each other and with other substances in the cell to govern the gene expression levels of mRNA and proteins which, in turn, determine the fun ...
s because of its capacity for fine regulation of promoters. In the absence of Tc or analogs likATc
basal expression of TetR-regulated promoters is low, but expression rises sharply in the presence of even a minute quantity of Tc. The ''tetA'' gene is also present in the widely used ''E. coli'' cloning vector
pBR322
pBR322 is a plasmid and was one of the first widely used '' E. coli'' cloning vectors. Created in 1977 in the laboratory of Herbert Boyer at the University of California, San Francisco, it was named after Francisco Bolivar Zapata, the postdoctora ...
, where it is often referred to by the name of its tetracycline-resistance phenotype, TetR, not to be confused with TetR.
Structure & Function
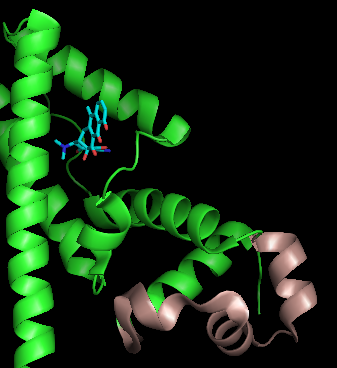 TetR functions as a
TetR functions as a homodimer
In biochemistry, a protein dimer is a macromolecular complex formed by two protein monomers, or single proteins, which are usually non-covalently bound. Many macromolecules, such as proteins or nucleic acids, form dimers. The word ''dimer'' has ...
. Each monomer consists of ten alpha helices
The alpha helix (α-helix) is a common motif in the secondary structure of proteins and is a right hand-helix conformation in which every backbone N−H group hydrogen bonds to the backbone C=O group of the amino acid located four residues earli ...
connected by loops and turns. The overall structure of TetR can be broken down into two DNA-binding domains (one per monomer) and a regulatory core, which is responsible for tetracycline recognition and dimerization. TetR dimerizes by making hydrophobic contacts within the regulatory core. There is a binding cavity for tetracycline in the outer helices of the regulatory domain. When tetracycline binds this cavity, it causes a conformational change that affects the DNA-binding domain so that TetR is no longer able to bind DNA. As a result, TetA and TetR are expressed. There is still some debate in the field whether tetracycline derivatives alone can cause this conformational change or whether tetracycline must be in complex with magnesium to bind TetR. (TetR typically binds tetracycline-Mg2+ complexes inside bacteria, but TetR binding to tetracycline alone has been observed in vitro.)
 The DNA-binding domains of TetR recognize a 15 base pair
The DNA-binding domains of TetR recognize a 15 base pair palindromic sequence
A palindromic sequence is a nucleic acid sequence in a double-stranded DNA or RNA molecule whereby reading in a certain direction (e.g. 5' to 3') on one strand is identical to the sequence in the same direction (e.g. 5' to 3') on the compleme ...
of the Tetmajor groove
Major (commandant in certain jurisdictions) is a military rank of commissioned officer status, with corresponding ranks existing in many military forces throughout the world. When used unhyphenated and in conjunction with no other indicators ...
s of the target DNA. Binding of TetR to its target DNA sequence causes changes in both the DNA and TetR. TetR causes widening of the major grooves as well as kinking of the DNA; one helix of the HTH motif of TetR adopts a 310 helical turn as the result of complex DNA interactions.
TetR Protein Family
 As of June 2005, this family of proteins had about 2,353 members that are transcriptional regulators. (Transcriptional regulators control gene expression.) These proteins contain a helix-turn-helix (HTH) motif that is the DNA-binding domain. The second helix is considered to be most important for DNA sequence specificity and often recognizes nucleic acids within the major groove of the double helix. In the majority of the family members, this motif is on the N-terminal end of the protein and is highly conserved. The high conservation of the HTH motif is not observed for the other domains of the protein. The differences observed in these other regulatory domains are likely due to differences in the molecules that each family member senses.
TetR protein family members are mostly transcriptional repressors, meaning that they prevent the expression of certain genes at the DNA level. These proteins can act on genes with various functions including antibiotic resistance, biosynthesis and metabolism, bacterial pathogenesis, and response to cell stress.
As of June 2005, this family of proteins had about 2,353 members that are transcriptional regulators. (Transcriptional regulators control gene expression.) These proteins contain a helix-turn-helix (HTH) motif that is the DNA-binding domain. The second helix is considered to be most important for DNA sequence specificity and often recognizes nucleic acids within the major groove of the double helix. In the majority of the family members, this motif is on the N-terminal end of the protein and is highly conserved. The high conservation of the HTH motif is not observed for the other domains of the protein. The differences observed in these other regulatory domains are likely due to differences in the molecules that each family member senses.
TetR protein family members are mostly transcriptional repressors, meaning that they prevent the expression of certain genes at the DNA level. These proteins can act on genes with various functions including antibiotic resistance, biosynthesis and metabolism, bacterial pathogenesis, and response to cell stress.
See also
* Tetracycline controlled transcriptional activationReferences
{{ReflistExternal links
Regulation of Antibiotic Resistance
Tetracycline antibiotics