Site-specific Recombinase Technology on:
[Wikipedia]
[Google]
[Amazon]
Site-specific recombinase technologies are
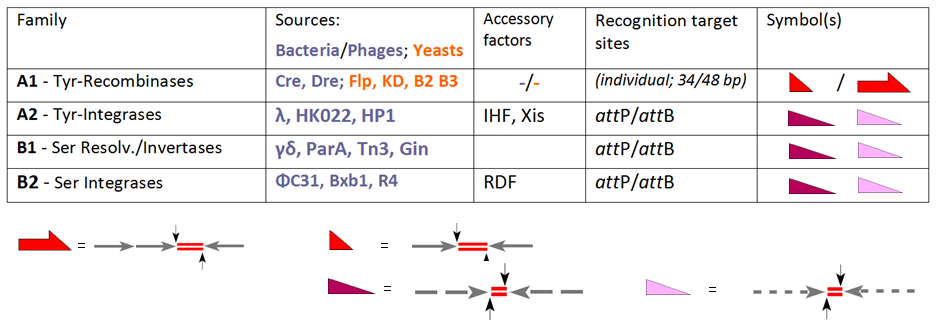 The founding member of the YR family is the lambda integrase, encoded by bacteriophage λ, enabling the integration of phage DNA into the bacterial genome. A common feature of this class is a conserved tyrosine nucleophile attacking the scissile DNA-phosphate to form a 3'-phosphotyrosine linkage. Early members of the SR family are closely related resolvase / DNA invertases from the bacterial transposons Tn3 and γδ, which rely on a catalytic serine responsible for attacking the scissile phosphate to form a 5'-phosphoserine linkage. These undisputed facts, however, were compromised by a good deal of confusion at the time other members entered the scene, for instance the YR recombinases Cre and Flp (capable of integration, excision/resolution as well as inversion), which were nevertheless welcomed as new members of the "integrase family". The converse examples are PhiC31 and related SRs, which were originally introduced as resolvase/invertases although, in the absence of auxiliary factors, integration is their only function. Nowadays the standard activity of each enzyme determines its classification reserving the general term "recombinase" for family members which, per se, comprise all three routes, INT, RES and INV:
Our table extends the selection of the conventional SSR systems and groups these according to their performance. All of these enzymes recombine two target sites, which are either identical (subfamily A1) or distinct (phage-derived enzymes in A2, B1 and B2). Whereas for A1 these sites have individual designations ("''FRT''" in case of Flp-recombinase, ''loxP'' for Cre-recombinase), the terms "''att''P" and "''att''B" (attachment sites on the phage and bacterial part, respectively) are valid in the other cases. In case of subfamily A1 we have to deal with short (usually 34 bp-) sites consisting of two (near-)identical 13 bp arms (arrows) flanking an 8 bp spacer (the crossover region, indicated by red line doublets). Note that for Flp there is an alternative, 48 bp site available with three arms, each accommodating a Flp unit (a so-called "protomer"). ''att''P- and ''att''B-sites follow similar architectural rules, but here the arms show only partial identity (indicated by the broken lines) and differ in both cases. These features account for relevant differences:
* recombination of two identical educt sites leads to product sites with the same composition, although they contain arms from both substrates; these conversions are reversible;
* in case of ''att''P x ''att''B recombination crossovers can only occur between these complementary partners in processes that lead to two different products (''att''P x ''att''B → ''att''R + ''att''L) in an irreversible fashion.
In order to streamline this chapter the following implementations will be focused on two recombinases (Flp and Cre) and just one integrase (PhiC31) since their spectrum covers the tools which, at present, are mostly used for directed genome modifications. This will be done in the framework of the following overview.
The founding member of the YR family is the lambda integrase, encoded by bacteriophage λ, enabling the integration of phage DNA into the bacterial genome. A common feature of this class is a conserved tyrosine nucleophile attacking the scissile DNA-phosphate to form a 3'-phosphotyrosine linkage. Early members of the SR family are closely related resolvase / DNA invertases from the bacterial transposons Tn3 and γδ, which rely on a catalytic serine responsible for attacking the scissile phosphate to form a 5'-phosphoserine linkage. These undisputed facts, however, were compromised by a good deal of confusion at the time other members entered the scene, for instance the YR recombinases Cre and Flp (capable of integration, excision/resolution as well as inversion), which were nevertheless welcomed as new members of the "integrase family". The converse examples are PhiC31 and related SRs, which were originally introduced as resolvase/invertases although, in the absence of auxiliary factors, integration is their only function. Nowadays the standard activity of each enzyme determines its classification reserving the general term "recombinase" for family members which, per se, comprise all three routes, INT, RES and INV:
Our table extends the selection of the conventional SSR systems and groups these according to their performance. All of these enzymes recombine two target sites, which are either identical (subfamily A1) or distinct (phage-derived enzymes in A2, B1 and B2). Whereas for A1 these sites have individual designations ("''FRT''" in case of Flp-recombinase, ''loxP'' for Cre-recombinase), the terms "''att''P" and "''att''B" (attachment sites on the phage and bacterial part, respectively) are valid in the other cases. In case of subfamily A1 we have to deal with short (usually 34 bp-) sites consisting of two (near-)identical 13 bp arms (arrows) flanking an 8 bp spacer (the crossover region, indicated by red line doublets). Note that for Flp there is an alternative, 48 bp site available with three arms, each accommodating a Flp unit (a so-called "protomer"). ''att''P- and ''att''B-sites follow similar architectural rules, but here the arms show only partial identity (indicated by the broken lines) and differ in both cases. These features account for relevant differences:
* recombination of two identical educt sites leads to product sites with the same composition, although they contain arms from both substrates; these conversions are reversible;
* in case of ''att''P x ''att''B recombination crossovers can only occur between these complementary partners in processes that lead to two different products (''att''P x ''att''B → ''att''R + ''att''L) in an irreversible fashion.
In order to streamline this chapter the following implementations will be focused on two recombinases (Flp and Cre) and just one integrase (PhiC31) since their spectrum covers the tools which, at present, are mostly used for directed genome modifications. This will be done in the framework of the following overview.

genome engineering
Genome editing, or genome engineering, or gene editing, is a type of genetic engineering in which DNA is inserted, deleted, modified or replaced in the genome of a living organism. Unlike early genetic engineering techniques that randomly inserts g ...
tools that depend on recombinase enzymes to replace targeted sections of DNA.
History
In the late 1980s gene targeting in murineembryonic stem cell
Embryonic stem cells (ESCs) are pluripotent stem cells derived from the inner cell mass of a blastocyst, an early-stage pre- implantation embryo. Human embryos reach the blastocyst stage 4–5 days post fertilization, at which time they consi ...
s (ESCs) enabled the transmission of mutations into the mouse germ line, and emerged as a novel option to study the genetic basis of regulatory networks as they exist in the genome. Still, classical gene targeting
Gene targeting (also, replacement strategy based on homologous recombination) is a genetic technique that uses homologous recombination to modify an endogenous gene. The method can be used to delete a gene, remove exons, add a gene and modify i ...
proved to be limited in several ways as gene functions became irreversibly destroyed by the marker gene that had to be introduced for selecting recombinant ESCs. These early steps led to animals in which the mutation was present in all cells of the body from the beginning leading to complex phenotypes and/or early lethality. There was a clear need for methods to restrict these mutations to specific points in development and specific cell types. This dream became reality when groups in the USA were able to introduce bacteriophage and yeast-derived site-specific recombination (SSR-) systems into mammalian cells as well as into the mouse.
Classification, properties and dedicated applications
Common genetic engineering strategies require a permanent modification of the target genome. To this end great sophistication has to be invested in the design of routes applied for the delivery of transgenes. Although for biotechnological purposes random integration is still common, it may result in unpredictable gene expression due to variable transgene copy numbers, lack of control about integration sites and associated mutations. The molecular requirements in the stem cell field are much more stringent. Here,homologous recombination
Homologous recombination is a type of genetic recombination in which genetic information is exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acids (usually DNA as in cellular organisms but may ...
(HR) can, in principle, provide specificity to the integration process, but for eukaryotes it is compromised by an extremely low efficiency. Although meganucleases, zinc-finger- and transcription activator-like effector nucleases (ZFNs and TALENs) are actual tools supporting HR, it was the availability of site-specific recombinases (SSRs) which triggered the rational construction of cell lines with predictable properties. Nowadays both technologies, HR and SSR can be combined in highly efficient "tag-and-exchange technologies".
Many site-specific recombination Site-specific recombination, also known as conservative site-specific recombination, is a type of genetic recombination in which DNA strand exchange takes place between segments possessing at least a certain degree of sequence homology. Enzymes kno ...
systems have been identified to perform these DNA rearrangements for a variety of purposes, but nearly all of these belong to either of two families, tyrosine recombinases (YR) and serine recombinases (SR), depending on their mechanism
Mechanism may refer to:
* Mechanism (engineering), rigid bodies connected by joints in order to accomplish a desired force and/or motion transmission
*Mechanism (biology), explaining how a feature is created
*Mechanism (philosophy), a theory that ...
. These two families can mediate up to three types of DNA rearrangements (integration, excision/resolution, and inversion) along different reaction routes based on their origin and architecture.
 The founding member of the YR family is the lambda integrase, encoded by bacteriophage λ, enabling the integration of phage DNA into the bacterial genome. A common feature of this class is a conserved tyrosine nucleophile attacking the scissile DNA-phosphate to form a 3'-phosphotyrosine linkage. Early members of the SR family are closely related resolvase / DNA invertases from the bacterial transposons Tn3 and γδ, which rely on a catalytic serine responsible for attacking the scissile phosphate to form a 5'-phosphoserine linkage. These undisputed facts, however, were compromised by a good deal of confusion at the time other members entered the scene, for instance the YR recombinases Cre and Flp (capable of integration, excision/resolution as well as inversion), which were nevertheless welcomed as new members of the "integrase family". The converse examples are PhiC31 and related SRs, which were originally introduced as resolvase/invertases although, in the absence of auxiliary factors, integration is their only function. Nowadays the standard activity of each enzyme determines its classification reserving the general term "recombinase" for family members which, per se, comprise all three routes, INT, RES and INV:
Our table extends the selection of the conventional SSR systems and groups these according to their performance. All of these enzymes recombine two target sites, which are either identical (subfamily A1) or distinct (phage-derived enzymes in A2, B1 and B2). Whereas for A1 these sites have individual designations ("''FRT''" in case of Flp-recombinase, ''loxP'' for Cre-recombinase), the terms "''att''P" and "''att''B" (attachment sites on the phage and bacterial part, respectively) are valid in the other cases. In case of subfamily A1 we have to deal with short (usually 34 bp-) sites consisting of two (near-)identical 13 bp arms (arrows) flanking an 8 bp spacer (the crossover region, indicated by red line doublets). Note that for Flp there is an alternative, 48 bp site available with three arms, each accommodating a Flp unit (a so-called "protomer"). ''att''P- and ''att''B-sites follow similar architectural rules, but here the arms show only partial identity (indicated by the broken lines) and differ in both cases. These features account for relevant differences:
* recombination of two identical educt sites leads to product sites with the same composition, although they contain arms from both substrates; these conversions are reversible;
* in case of ''att''P x ''att''B recombination crossovers can only occur between these complementary partners in processes that lead to two different products (''att''P x ''att''B → ''att''R + ''att''L) in an irreversible fashion.
In order to streamline this chapter the following implementations will be focused on two recombinases (Flp and Cre) and just one integrase (PhiC31) since their spectrum covers the tools which, at present, are mostly used for directed genome modifications. This will be done in the framework of the following overview.
The founding member of the YR family is the lambda integrase, encoded by bacteriophage λ, enabling the integration of phage DNA into the bacterial genome. A common feature of this class is a conserved tyrosine nucleophile attacking the scissile DNA-phosphate to form a 3'-phosphotyrosine linkage. Early members of the SR family are closely related resolvase / DNA invertases from the bacterial transposons Tn3 and γδ, which rely on a catalytic serine responsible for attacking the scissile phosphate to form a 5'-phosphoserine linkage. These undisputed facts, however, were compromised by a good deal of confusion at the time other members entered the scene, for instance the YR recombinases Cre and Flp (capable of integration, excision/resolution as well as inversion), which were nevertheless welcomed as new members of the "integrase family". The converse examples are PhiC31 and related SRs, which were originally introduced as resolvase/invertases although, in the absence of auxiliary factors, integration is their only function. Nowadays the standard activity of each enzyme determines its classification reserving the general term "recombinase" for family members which, per se, comprise all three routes, INT, RES and INV:
Our table extends the selection of the conventional SSR systems and groups these according to their performance. All of these enzymes recombine two target sites, which are either identical (subfamily A1) or distinct (phage-derived enzymes in A2, B1 and B2). Whereas for A1 these sites have individual designations ("''FRT''" in case of Flp-recombinase, ''loxP'' for Cre-recombinase), the terms "''att''P" and "''att''B" (attachment sites on the phage and bacterial part, respectively) are valid in the other cases. In case of subfamily A1 we have to deal with short (usually 34 bp-) sites consisting of two (near-)identical 13 bp arms (arrows) flanking an 8 bp spacer (the crossover region, indicated by red line doublets). Note that for Flp there is an alternative, 48 bp site available with three arms, each accommodating a Flp unit (a so-called "protomer"). ''att''P- and ''att''B-sites follow similar architectural rules, but here the arms show only partial identity (indicated by the broken lines) and differ in both cases. These features account for relevant differences:
* recombination of two identical educt sites leads to product sites with the same composition, although they contain arms from both substrates; these conversions are reversible;
* in case of ''att''P x ''att''B recombination crossovers can only occur between these complementary partners in processes that lead to two different products (''att''P x ''att''B → ''att''R + ''att''L) in an irreversible fashion.
In order to streamline this chapter the following implementations will be focused on two recombinases (Flp and Cre) and just one integrase (PhiC31) since their spectrum covers the tools which, at present, are mostly used for directed genome modifications. This will be done in the framework of the following overview.

Reaction routes
The mode integration/resolution and inversion (INT/RES and INV) depend on the orientation of recombinase target sites (RTS), among these pairs of ''att''P and ''att''B. Section C indicates, in a streamlined fashion, the wayrecombinase-mediated cassette exchange RMCE (recombinase-mediated cassette exchange) is a procedure in reverse genetics allowing the systematic, repeated modification of higher eukaryotic genomes by targeted integration, based on the features of site-specific recombination processes (SS ...
(RMCE) can be reached by synchronous double-reciprocal crossovers (rather than integration, followed by resolution).
Tyr-Recombinases are reversible, while the Ser-Integrase is unidirectional. Of note is the way reversible Flp (a Tyr recombinase) integration/resolution is modulated by 48 bp (in place of 34 bp minimal) ''FRT'' versions: the extra 13 bp arm serves as a Flp "landing path" contributing to the formation of the synaptic complex, both in the context of Flp-INT and Flp-RMCE functions (see the respective equilibrium situations). While it is barely possible to prevent the (entropy-driven) reversion of integration in section A for Cre and hard to achieve for Flp, RMCE can be completed if the donor plasmid is provided at an excess due to the bimolecular character of both the forward- and the reverse reaction. Posing both ''FRT'' sites in an inverse manner will lead to an equilibrium of both orientations for the insert (green arrow). In contrast to Flp, the Ser integrase PhiC31 (bottom representations) leads to unidirectional integration, at least in the absence of an recombinase-directionality (RDF-)factor. Relative to Flp-RMCE, which requires two different ("heterospecific") ''FRT''-spacer mutants, the reaction partner (''att''B) of the first reacting ''att''P site is hit arbitrarily, such that there is no control over the direction the donor cassette enters the target (cf. the alternative products). Also different from Flp-RMCE, several distinct RMCE targets cannot be mounted in parallel, owing to the lack of heterospecific (non-crossinteracting) ''att''P/''att''B combinations.
Cre recombinase
Cre recombinase
Cre recombinase is a tyrosine recombinase enzyme derived from the P1 bacteriophage. The enzyme uses a topoisomerase I-like mechanism to carry out site specific recombination events. The enzyme (38kDa) is a member of the integrase family of site ...
(Cre) is able to recombine specific sequences of DNA without the need for cofactors
Cofactor may also refer to:
* Cofactor (biochemistry), a substance that needs to be present in addition to an enzyme for a certain reaction to be catalysed
* A domain parameter in elliptic curve cryptography, defined as the ratio between the order ...
. The enzyme recognizes 34 base pair DNA sequences called ''loxP'' ("locus of crossover in phage P1"). Depending on the orientation of target sites with respect to one another, Cre will integrate/excise or invert DNA sequences. Upon the excision (called "resolution" in case of a circular substrate) of a particular DNA region, normal gene expression is considerably compromised or terminated.
Due to the pronounced resolution activity of Cre, one of its initial applications was the excision of ''lox''P-flanked ("floxed") genes leading to cell-specific gene knockout of such a floxed gene after Cre becomes expressed in the tissue of interest. Current technologies incorporate methods, which allow for both the spatial and temporal control of Cre activity. A common method facilitating the spatial control of genetic alteration involves the selection of a tissue-specific promoter to drive Cre expression. Placement of Cre under control of such a promoter results in localized, tissue-specific expression. As an example, Leone et al. have placed the transcription unit under the control of the regulatory sequences of the myelin
Myelin is a lipid-rich material that surrounds nerve cell axons (the nervous system's "wires") to insulate them and increase the rate at which electrical impulses (called action potentials) are passed along the axon. The myelinated axon can be ...
proteolipid protein (PLP) gene, leading to induced removal of targeted gene sequences in oligodendrocytes
Oligodendrocytes (), or oligodendroglia, are a type of neuroglia whose main functions are to provide support and insulation to axons in the central nervous system of jawed vertebrates, equivalent to the function performed by Schwann cells in the ...
and Schwann cells
Schwann cells or neurolemmocytes (named after German physiologist Theodor Schwann) are the principal glia of the peripheral nervous system (PNS). Glial cells function to support neurons and in the PNS, also include satellite cells, olfactory en ...
. The specific DNA fragment recognized by Cre remains intact in cells, which do not express the PLP gene; this in turn facilitates empirical observation of the localized effects of genome alterations in the myelin sheath that surround nerve fibers in the central nervous system
The central nervous system (CNS) is the part of the nervous system consisting primarily of the brain and spinal cord. The CNS is so named because the brain integrates the received information and coordinates and influences the activity of all par ...
(CNS) and the peripheral nervous system
The peripheral nervous system (PNS) is one of two components that make up the nervous system of bilateral animals, with the other part being the central nervous system (CNS). The PNS consists of nerves and ganglia, which lie outside the brain ...
(PNS). Selective Cre expression has been achieved in many other cell types and tissues as well.
In order to control temporal activity of the excision reaction, forms of Cre which take advantage of various ligand
In coordination chemistry, a ligand is an ion or molecule (functional group) that binds to a central metal atom to form a coordination complex. The bonding with the metal generally involves formal donation of one or more of the ligand's electr ...
binding domains have been developed. One successful strategy for inducing specific temporal Cre activity involves fusing the enzyme with a mutated ligand-binding domain for the human estrogen receptor
Estrogen receptors (ERs) are a group of proteins found inside cells. They are receptors that are activated by the hormone estrogen ( 17β-estradiol). Two classes of ER exist: nuclear estrogen receptors (ERα and ERβ), which are members of the ...
(ERt). Upon the introduction of tamoxifen
Tamoxifen, sold under the brand name Nolvadex among others, is a selective estrogen receptor modulator used to prevent breast cancer in women and treat breast cancer in women and men. It is also being studied for other types of cancer. It has b ...
(an estrogen receptor antagonist
A receptor antagonist is a type of receptor ligand or drug that blocks or dampens a biological response by binding to and blocking a receptor rather than activating it like an agonist. Antagonist drugs interfere in the natural operation of rece ...
), the Cre-ERt construct is able to penetrate the nucleus and induce targeted mutation. ERt binds tamoxifen with greater affinity than endogenous
Endogenous substances and processes are those that originate from within a living system such as an organism, tissue, or cell.
In contrast, exogenous substances and processes are those that originate from outside of an organism.
For example, es ...
estrogens
Estrogen or oestrogen is a category of sex hormone responsible for the development and regulation of the female reproductive system and secondary sex characteristics. There are three major endogenous estrogens that have estrogenic hormonal acti ...
, which allows Cre-ERt to remain cytoplasmic
In cell biology, the cytoplasm is all of the material within a eukaryotic cell, enclosed by the cell membrane, except for the cell nucleus. The material inside the nucleus and contained within the nuclear membrane is termed the nucleoplasm. Th ...
in animals untreated with tamoxifen. The temporal control of SSR activity by tamoxifen permits genetic changes to be induced later in embryogenesis
An embryo is an initial stage of development of a multicellular organism. In organisms that reproduce sexually, embryonic development is the part of the life cycle that begins just after fertilization of the female egg cell by the male sperm ...
and/or in adult tissues. This allows researchers to bypass embryonic lethality while still investigating the function of targeted genes.
Recent extensions of these general concepts led to generating the "Cre-zoo", i.e. collections of hundreds of mouse strains for which defined genes can be deleted by targeted Cre expression.
Flp recombinase
In its natural host (S. cerevisiae) the Flp/''FRT'' system enables replication of a "2μ plasmid" by the inversion of a segment that is flanked by two identical, but oppositely oriented ''FRT'' sites ("flippase" activity). This inversion changes the relative orientation of replication forks within the plasmid enabling "rolling circle"—amplification of the circular 2μ entity before the multimeric intermediates are resolved to release multiple monomeric products. Whereas 34 bp minimal ''FRT'' sites favor excision/resolution to a similar extent as the analogue ''lox''P sites for Cre, the natural, more extended 48 bp FRT variants enable a higher degree of integration, while overcoming certain promiscuous interactions as described for phage enzymes like Cre- and PhiC31. An additional advantage is the fact, that simple rules can be applied to generate heterospecific ''FRT'' sites which undergo crossovers with equal partners but nor with wild type ''FRT''s. These facts have enabled, since 1994, the development and continuous refinements ofrecombinase-mediated cassette exchange RMCE (recombinase-mediated cassette exchange) is a procedure in reverse genetics allowing the systematic, repeated modification of higher eukaryotic genomes by targeted integration, based on the features of site-specific recombination processes (SS ...
(RMCE-)strategies permitting the clean exchange of a target cassette for an incoming donor cassette.
Based on the RMCE technology, a particular resource of pre-characterized ES-strains that lends itself to further elaboration has evolved in the framework of the EUCOMM (European Conditional Mouse Mutagenesis) program, based on the now established Cre- and/or Flp-based "FlExing" (Flp-mediated excision/inversion) setups, involving the excision and inversion activities. Initiated in 2005, this project focused first on saturation mutagenesis to enable complete functional annotation of the mouse genome (coordinated by the International Knockout-Mouse Consortium, IKMC) with the ultimate goal to have all protein genes mutated via gene trapping and -targeting in murine ES cells. These efforts mark the top of various "tag-and-exchange" strategies, which are dedicated to tagging a distinct genomic site such that the "tag" can serve as an address to introduce novel (or alter existing) genetic information. The tagging step ''per se'' may address certain classes of integration sites by exploiting integration preferences of retroviruses or even site specific integrases like PhiC31, both of which act in an essentially unidirectional fashion.
The traditional, laborious "tag-and-exchange" procedures relied on two successive homologous recombination (HR-)steps, the first one ("HR1") to introduce a tag consisting of a selection marker gene. "HR2" was then used to replace the marker by the "GOI. In the first ("knock-out"-) reaction the gene was tagged with a selectable marker, typically by insertion of a hygtk ( /- cassette providing G418 resistance. In the following "knock-in" step, the tagged genomic sequence was replaced by homologous genomic sequences with certain mutations. Cell clones could then be isolated by their resistance to ganciclovir due to loss of the HSV-tk gene, i.e. ("negative selection"). This conventional two-step tag-and-exchange procedure could be streamlined after the advent of RMCE, which could take over and add efficiency to the knock-in step.
PhiC31 integrase
Without much doubt, Serintegrase
Retroviral integrase (IN) is an enzyme produced by a retrovirus (such as HIV) that integrates—forms covalent links between—its genetic information into that of the host cell it infects. Retroviral INs are not to be confused with phage int ...
s are the current tools of choice for integrating transgenes into a restricted number of well-understood genomic acceptor sites that mostly (but not always) mimic the phage ''att''P site in that they attract an ''att''B-containing donor vector. At this time the most prominent member is PhiC31-INT with proven potential in the context of human and mouse genomes.
Contrary to the above Tyr recombinases, PhiC31-INT as such acts in a unidirectional manner, firmly locking in the donor vector at a genomically anchored target. An obvious advantage of this system is that it can rely on unmodified, native ''att''P (acceptor) and ''att''B donor sites. Additional benefits (together with certain complications) may arise from the fact that mouse and human genomes per se contain a limited number of endogenous targets (so called "''att''P-pseudosites"). Available information suggests that considerable DNA sequence requirements let the integrase recognize fewer sites than retroviral or even transposase-based integration systems opening its career as a superior carrier vehicle for the transport and insertion at a number of well established genomic sites, some of which with so called "safe-harbor" properties.
Exploiting the fact of specific (''att''P x ''att''B) recombination routes, RMCE becomes possible without requirements for synthetic, heterospecific ''att''-sites. This obvious advantage, however comes at the expense of certain shortcomings, such as lack of control about the kind or directionality of the entering (donor-) cassette. Further restrictions are imposed by the fact that irreversibility does not permit standard multiplexing-RMCE setups including "serial RMCE" reactions, i.e., repeated cassette exchanges at a given genomic ''locus''.
Outlook and perspectives
Annotation of the human and mouse genomes has led to the identification of >20 000 protein-coding genes and >3 000 noncoding RNA genes, which guide the development of the organism from fertilization through embryogenesis to adult life. Although dramatic progress is noted, the relevance of rare gene variants has remained a central topic of research. As one of the most important platforms for dealing with vertebrate gene functions on a large scale, genome-wide genetic resources of mutant murine ES cells have been established. To this end four international programs aimed at saturation mutagenesis of the mouse genome have been founded in Europe and North America (EUCOMM, KOMP, NorCOMM, and TIGM). Coordinated by the International Knockout Mouse Consortium (IKSC) these ES-cell repositories are available for exchange between international research units. Present resources comprise mutations in 11 539 unique genes, 4 414 of these conditional. The relevant technologies have now reached a level permitting their extension to other mammalian species and to human stem cells, most prominently those with an iPS (induced pluripotent) status.See also
*Site-specific recombination Site-specific recombination, also known as conservative site-specific recombination, is a type of genetic recombination in which DNA strand exchange takes place between segments possessing at least a certain degree of sequence homology. Enzymes kno ...
* Recombinase-mediated cassette exchange RMCE (recombinase-mediated cassette exchange) is a procedure in reverse genetics allowing the systematic, repeated modification of higher eukaryotic genomes by targeted integration, based on the features of site-specific recombination processes (SS ...
* Cre recombinase
Cre recombinase is a tyrosine recombinase enzyme derived from the P1 bacteriophage. The enzyme uses a topoisomerase I-like mechanism to carry out site specific recombination events. The enzyme (38kDa) is a member of the integrase family of site ...
* Cre-Lox recombination
Cre-Lox recombination is a site-specific recombinase technology, used to carry out deletions, insertions, translocations and inversions at specific sites in the DNA of cells. It allows the DNA modification to be targeted to a specific cell type ...
* FLP-FRT recombination
In genetics, Flp-''FRT'' recombination is a site-directed recombination technology, increasingly used to manipulate an organism's DNA under controlled conditions ''in vivo''. It is analogous to Cre-''lox'' recombination but involves the recombi ...
* Genetic recombination
Genetic recombination (also known as genetic reshuffling) is the exchange of genetic material between different organisms which leads to production of offspring with combinations of traits that differ from those found in either parent. In eukaryo ...
* Homologous recombination
Homologous recombination is a type of genetic recombination in which genetic information is exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acids (usually DNA as in cellular organisms but may ...
References
External links
*http://www.knockoutmouse.org/ *{{cite journal , last1 = Emes , first1 = RD , last2 = Goodstadt , first2 = L , last3 = Winter , first3 = EE , last4 = Ponting , first4 = CP , year = 2003 , title = Comparison of the genomes of human and mouse lays the foundation of genome zoology , journal = Hum Mol Genet , volume = 12 , issue = 7, pages = 701–9 , doi=10.1093/hmg/ddg078 , pmid=12651866, doi-access = free Genetic engineering