Shavenbaby Locus on:
[Wikipedia]
[Google]
[Amazon]
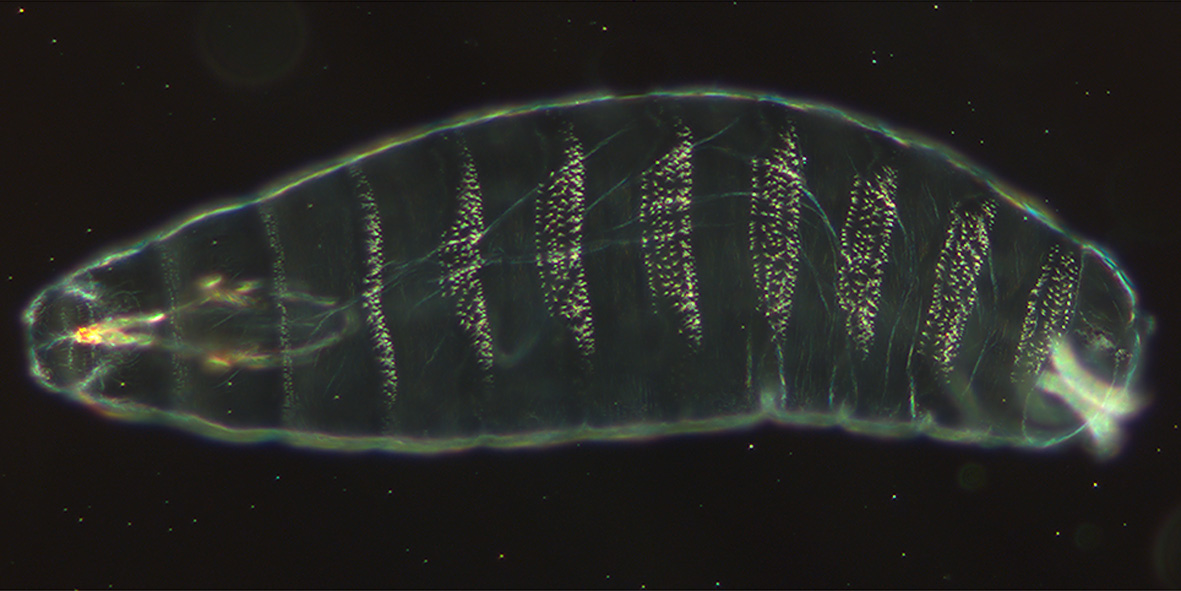 Trichomes likely serve a variety of purposes. In larvae, trichomes likely help with larval locomotion. By alternating between bands of trichomes and naked cuticle, larvae can tread across different surfaces. Additionally, trichomes may contribute to hydrophobicity and even stabilize adult flight.
Trichomes likely serve a variety of purposes. In larvae, trichomes likely help with larval locomotion. By alternating between bands of trichomes and naked cuticle, larvae can tread across different surfaces. Additionally, trichomes may contribute to hydrophobicity and even stabilize adult flight.

 Enhancer redundancy is a commonly observed phenomenon. Why would evolution evolve redundant enhancers? The mystery of enhancer redundancy was partially resolved by studying the ''shavenbaby'' locus in 2010. Frankel et al. found that the redundant enhancers help maintain proper ''shavenbaby'' expression under different temperature stresses, canalizing its expression. This finding was also observed eight years later for redundant mammalian enhancers, suggesting that this observation is not limited to ''Drosophila''. Redundant enhancers have also been observed to use different
Enhancer redundancy is a commonly observed phenomenon. Why would evolution evolve redundant enhancers? The mystery of enhancer redundancy was partially resolved by studying the ''shavenbaby'' locus in 2010. Frankel et al. found that the redundant enhancers help maintain proper ''shavenbaby'' expression under different temperature stresses, canalizing its expression. This finding was also observed eight years later for redundant mammalian enhancers, suggesting that this observation is not limited to ''Drosophila''. Redundant enhancers have also been observed to use different
 ''E3N'' was first described in Crocker et al., 2015, and was found to encode "homotypic clusters" of binding sites for the
''E3N'' was first described in Crocker et al., 2015, and was found to encode "homotypic clusters" of binding sites for the
Overview of ''shavenbaby (svb)''
The ''shavenbaby'' (''svb'') or ''ovo'' gene encodes atranscription factor
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The fu ...
in ''Drosophila
''Drosophila'' () is a genus of flies, belonging to the family Drosophilidae, whose members are often called "small fruit flies" or (less frequently) pomace flies, vinegar flies, or wine flies, a reference to the characteristic of many species ...
'' responsible for inducing cells to become hair-like projections called trichomes or microtrichia. Many of the major developmental signaling pathways converge at the ''shavenbaby'' locus, which then regulates over 150 downstream target genes. The "hourglass" shape of this gene regulatory network makes ''shavenbaby'' the master regulator
In genetics, a master regulator is a gene at the top of a gene regulation hierarchy, particularly in regulatory pathways related to cell fate and differentiation.
Examples
Most genes considered master regulators code for transcription factor prot ...
of trichome formation. The unique setup of the gene regulatory network made trichomes an excellent readout to identify important developmental genes during the forward genetics Heidelberg Screen. Additionally, ''shavenbaby'' is considered to be an "evolutionary hotspot", and experiments have shown that changes in this gene cause the loss of dorsal cuticular hairs in ''Drosophila sechellia
''Drosophila sechellia'' is a species of fruit fly, used in lab studies of speciation because it can mate with ''Drosophila simulans''.
''Drosophila sechellia'' is endemic to (some of) the Seychelles, and was one of 12 fruit fly genomes sequenc ...
'' larvae.
 Trichomes likely serve a variety of purposes. In larvae, trichomes likely help with larval locomotion. By alternating between bands of trichomes and naked cuticle, larvae can tread across different surfaces. Additionally, trichomes may contribute to hydrophobicity and even stabilize adult flight.
Trichomes likely serve a variety of purposes. In larvae, trichomes likely help with larval locomotion. By alternating between bands of trichomes and naked cuticle, larvae can tread across different surfaces. Additionally, trichomes may contribute to hydrophobicity and even stabilize adult flight.
Transcriptional inputs for ''svb''
The ''shavenbaby'' locus is regulated by multiple signaling pathways, including the HOX factors,Wingless
The Wnt signaling pathways are a group of signal transduction pathways which begin with proteins that pass signals into a cell through cell surface receptors. The name Wnt is a portmanteau created from the names Wingless and Int-1. Wnt signaling p ...
, EGF-R, Hedgehog
A hedgehog is a spiny mammal of the subfamily Erinaceinae, in the eulipotyphlan family Erinaceidae. There are seventeen species of hedgehog in five genera found throughout parts of Europe, Asia, and Africa, and in New Zealand by introducti ...
, and Notch signaling. Additionally, the transcription factors SoxNeuro, Pointed, and Dichaete regulate ''shavenbaby'' expression.
Engrailed and Hedgehog activate EGFR
During stage 12 of embryonic development, Engrailed is expressed in a subset of cells, which activates the hedgehog signaling pathway. The Hedgehog signal is received by cells expressing Patched, which induces expression of ''rhomboid'' (''rho'') with Serrate-Notch signaling, which activates the EGFR signaling pathway. The drosophila EGF receptor (DER) is responsible for activating ''shavenbaby'' both directly and by driving expression of the factors ''SoxNeuro'' and ''Dichaete''. Other transcription factors such as Ultrabithorax and its cofactor Homothorax also interact with the different ''shavenbaby'' enhancers to activate expression.Wingless signaling represses ''shavenbaby''
During stage 12, the Hedgehog signaling pathway induces expression of the Wingless signal. The Wingless signaling pathway is responsible for repressing ''shavenbaby'' activity, and cells expressing Wingless have naked cuticle. Furthermore, mutations to the Wingless gene produce a lawn of trichomes in the naked region. Wingless signaling has been characterized to specifically integrate at the ''shavenbaby'' ''E3'' enhancer, which also produces a lawn of expression in Wingless mutants. Wingless signaling is repressed by both SoxNeuro and Dichaete, products of the EGFR signaling pathway.Developmental enhancers of ''svb''
Developmentalenhancers
In genetics, an enhancer is a short (50–1500 bp) region of DNA that can be bound by proteins ( activators) to increase the likelihood that transcription of a particular gene will occur. These proteins are usually referred to as transcription ...
are DNA sequences which control the spatial-temporal patterning of genes during development to set up the bodyplan of an organism. Developmental enhancers are thought to be the main drivers of phenotypic evolution. There are currently seven putative developmental enhancers in the ''shavenbaby'' locus: ''DG2, DG3, Z1.3, A, E3, E6,'' and ''7H''. All of these enhancers are pleiotropic
Pleiotropy (from Greek , 'more', and , 'way') occurs when one gene influences two or more seemingly unrelated phenotypic traits. Such a gene that exhibits multiple phenotypic expression is called a pleiotropic gene. Mutation in a pleiotropic g ...
, expressing ''shavenbaby'' across different developmental stages. The enhancers are somewhat modular, where different patterning components are partitioned to different enhancers. However, many of the expression patterns overlap with each other making the enhancers seemingly redundant.
 Enhancer redundancy is a commonly observed phenomenon. Why would evolution evolve redundant enhancers? The mystery of enhancer redundancy was partially resolved by studying the ''shavenbaby'' locus in 2010. Frankel et al. found that the redundant enhancers help maintain proper ''shavenbaby'' expression under different temperature stresses, canalizing its expression. This finding was also observed eight years later for redundant mammalian enhancers, suggesting that this observation is not limited to ''Drosophila''. Redundant enhancers have also been observed to use different
Enhancer redundancy is a commonly observed phenomenon. Why would evolution evolve redundant enhancers? The mystery of enhancer redundancy was partially resolved by studying the ''shavenbaby'' locus in 2010. Frankel et al. found that the redundant enhancers help maintain proper ''shavenbaby'' expression under different temperature stresses, canalizing its expression. This finding was also observed eight years later for redundant mammalian enhancers, suggesting that this observation is not limited to ''Drosophila''. Redundant enhancers have also been observed to use different transcription factor
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The fu ...
s, incorporating a diverse set of signaling inputs to canalize gene expression under different environmental stresses.
The ''E3'' enhancer
The ''E3'' enhancer is a 1,042 base-pair (bp) enhancer which drives ''shavenbaby'' on the ventral side of stages 15 and 16+ embryos and larvae. ''E3'' is also expressed pleiotropically in the pharynx and esophagus or third-instar larvae. In adult ''Drosophila, E3'' is expressed in the abdomen, head, legs, and wing. The ''E3'' fragment has been tested as smaller fragments such as ''E3-14'' and ''E3N''. Unlike the other ''shavenbaby'' enhancers, ''E3'' activity is maintained in ''Drosophila sechellia''. ''E3N'' was first described in Crocker et al., 2015, and was found to encode "homotypic clusters" of binding sites for the
''E3N'' was first described in Crocker et al., 2015, and was found to encode "homotypic clusters" of binding sites for the transcription factor
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The fu ...
: Ultrabithorax
Ultrabithorax (Ubx) is a homeobox gene found in insects, and is used in the regulation of patterning in morphogenesis. There are many possible products of this gene, which function as transcription factors. Ubx is used in the specification of se ...
(Ubx). These binding sites, however, were non-canonical, and Ubx binds to ''E3N'' at a very low-affinity. Mutations to increase the affinity of these binding sites caused the ectopic binding of other Homeobox
A homeobox is a DNA sequence, around 180 base pairs long, that regulates large-scale anatomical features in the early stages of embryonic development. For instance, mutations in a homeobox may change large-scale anatomical features of the full- ...
(HOX) factors, resulting in ectopic enhancer expression. HOX factors license the identity of cells, locking them into a fate to produce a particular structure such as wings, halteres, antennae, abdomen, etc. All of the HOX factors are evolutionarily related, and bind to the same homeodomain sequence: TAAT. How enhancers encode the specific binding of certain HOX factors and prevent the ectopic binding of others is called the "Hox Paradox". The ''E3N'' study from Crocker et al., 2015 provided an answer to the "Hox Paradox", by suggesting that low-affinity binding sites would provide the specificity, and encoding clusters of the sites would account for the potential weak activation. Low-affinity transcription factor binding sites have also been observed in other enhancers.
In a follow-up study, Fuqua et al. created a library of random mutants to the ''E3N'' enhancer to study the enhancer grammar and how enhancers can evolve. The study revealed that even single point mutations had a significant effect on the enhancer expression pattern. Furthermore, the mutations affected multiple components of the pattern. This pleiotropic nature of the mutations was demonstrated when the emergence of novel salivary gland or mouth hook expression was linked with the nearly complete loss of the original embryonic expression pattern. Additionally, changes to the low-affinity Ultrabithorax binding sites resulted in pleiotropic effects modulating the timing
Timing is the tracking or planning of the spacing of events in time. It may refer to:
* Timekeeping, the process of measuring the passage of time
* Synchronization, controlling the timing of a process relative to another process
* Time metrolog ...
, pattern intensity, and ectopic expression. The authors concluded that enhancers are densely encoded with regulatory information and enhancer mutations are usually pleiotropic. Other recent studies in the ''yellow spot'' enhancer and the Sonic Hedgehog
Sonic hedgehog protein (SHH) is encoded for by the ''SHH'' gene. The protein is named after the character ''Sonic the Hedgehog''.
This signaling molecule is key in regulating embryonic morphogenesis in all animals. SHH controls organogenesis and ...
''ZRS'' enhancer also support this claim. These findings may even suggest that the underlying cis-regulatory logic of an enhancer may constrain its evolution, a claim also made my Preger Ben-Noon et al.
The ''E6'' enhancer
The ''E6'' enhancer is expressed in the dorsal and quaternary cells of ''Drosophila'' embryos, larvae, and in the pupal epidermis. The ''E6'' enhancer is one of the five enhancers that contributed to the loss of the larval dorsal trichomes in ''Drosophila sechellia''. The molecular mechanism for this loss of expression was resolved by Preger Ben-Noon et al., where ''sechellia-E6'' consecutively accumulated mutations in activator sites for Arrowhead and Pannier and gained a binding site for the repressor Abrupt. These mutations contributed to a 46% decrease in total embryonic ''shavenbaby'' expression, and affected the pleiotropic expression in the pupal epidermis.The ''Z1.3'' enhancer
The ''Z1.3'' enhancer is a minimalized fragment of the ''Z'' enhancer, and drives expression in the embryonic quaternary cells, the larval pharynx and proventriculus, and the pupal epidermis. The ''Z1.3'' enhancer contributed to an estimated 28% loss of total embryonic expression in ''Drosophila sechellia.'' However, unlike in ''E6,'' the mutations that affected the embryonic pattern of ''Z1.3'' had no effect on its pleiotropic pupal epidermis expression. Preger Ben-Noon et al. further dissected the ''Z1.3'' enhancer and were able to minimalize the pleiotropic activity into two separate enhancers: ''Z0.3'' and ''Z1.3R''.The ''DG3'' enhancer
The ''DG3'' enhancer is primarily expressed in the ventral embryonic epidermis along with ''E3N'' and ''7H.'' In larvae, ''DG3'' is expressed in the dorsal and ventral regions, in the pharynix, esophagus, and proventriculus, and in the pupal epidermis. A closer look at the ventral nuclei reveals that the ''shavenbaby'' gene physically colocalizes with higher concentrations of the Ultrabithorax protein and its cofactor Homothorax. Additionally, the ''Drosophila'' line ''Df(svb)108'' contains a deletion in the ''DG2, DG3, and Z'' enhancers. Heat shocking these lines does induce a slight decrease in the number of ventral trichomes. A closer look at the nuclei of these individual cells reveals both lower quanitifiable levels of the ''shavenbaby'' transcript and weaker nuclear microenvironment interactions between the ventral enhancers . Interestingly, transcript levels and the microenvironment can be stabilized by crossing flies carrying the deletion with flies carrying an artificial BAC of the ''shavenbaby'' locus. The studies from Tsai et al. reveals microenvironments and potentially transvection to be potential mechanisms for how redundant enhancers canalize gene expression.The ''7H'' enhancer
The ''7H'' enhancer drives expression in both the ventral and dorsal embryonic and larval epidermis, the larval pharynx, and the pupal epidermis. Deletion of the ''7H'' enhancer results in a 38% decrease in total embryonic ''shavenbaby'' expression. ''7H, DG3,'' and ''E3N'' are the primary ventral enhancers in the embryo.Trichome formation
Shavenbaby activates over 150 different downstream targets to express actin-remodeling proteins to form the denticle. Some of these factors include ''forked, shavenoid, singed, wasp,'' yellow, and ''miniature''. Activation of these target genes is also dependent on SoxNeuro, one of the regulators of ''shavenbaby.'' Together, SoxNeuro and Shavenbaby act cooperatively to shape the denticles.References
{{reflist Drosophila melanogaster genes