Reprogramming on:
[Wikipedia]
[Google]
[Amazon]
In biology, reprogramming refers to erasure and remodeling of
 The mouse
The mouse  After fertilization some cells of the newly formed embryo migrate to the germinal ridge and will eventually become the
After fertilization some cells of the newly formed embryo migrate to the germinal ridge and will eventually become the
 The Figure in this section indicates the central roles of ten-eleven translocation methylcytosine dioxygenases (TETs) in the demethylation of 5-methylcytosine to form cytosine. As reviewed in 2018, 5mC is very often initially oxidized by TET dioxygenases to generate 5-hydroxymethylcytosine (5hmC). In successive steps (see Figure) TET enzymes further hydroxylate 5hmC to generate 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC). Thymine-DNA glycosylase (TDG) recognizes the intermediate bases 5fC and 5caC and excises the
The Figure in this section indicates the central roles of ten-eleven translocation methylcytosine dioxygenases (TETs) in the demethylation of 5-methylcytosine to form cytosine. As reviewed in 2018, 5mC is very often initially oxidized by TET dioxygenases to generate 5-hydroxymethylcytosine (5hmC). In successive steps (see Figure) TET enzymes further hydroxylate 5hmC to generate 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC). Thymine-DNA glycosylase (TDG) recognizes the intermediate bases 5fC and 5caC and excises the
 For a TET enzyme to initiate demethylation it must first be recruited to a methylated CpG site in DNA. Two of the proteins shown to recruit a TET enzyme to a methylated cytosine in DNA are OGG1 (see figure Initiation of DNA demthylation) and EGR1.
For a TET enzyme to initiate demethylation it must first be recruited to a methylated CpG site in DNA. Two of the proteins shown to recruit a TET enzyme to a methylated cytosine in DNA are OGG1 (see figure Initiation of DNA demthylation) and EGR1.
 An
An

 Unlike nuclear transfer and cell fusion, defined factors do not require a full genome, only reprogramming factors. These reprogramming factors include
Unlike nuclear transfer and cell fusion, defined factors do not require a full genome, only reprogramming factors. These reprogramming factors include
epigenetic
In biology, epigenetics is the study of changes in gene expression that happen without changes to the DNA sequence. The Greek prefix ''epi-'' (ἐπι- "over, outside of, around") in ''epigenetics'' implies features that are "on top of" or "in ...
marks, such as DNA methylation
DNA methylation is a biological process by which methyl groups are added to the DNA molecule. Methylation can change the activity of a DNA segment without changing the sequence. When located in a gene promoter (genetics), promoter, DNA methylati ...
, during mammalian development or in cell culture. Such control is also often associated with alternative covalent modifications of histones
In biology, histones are highly Base (chemistry), basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei and in most Archaea, Archaeal Phylum, phyla. They act as spools around which DNA winds to create st ...
.
Reprogrammings that are both large scale (10% to 100% of epigenetic marks) and rapid (hours to a few days) occur at three life stages of mammals. Almost 100% of epigenetic marks are reprogrammed in two short periods early in development after fertilization
Fertilisation or fertilization (see American and British English spelling differences#-ise, -ize (-isation, -ization), spelling differences), also known as generative fertilisation, syngamy and impregnation, is the fusion of gametes to give ...
of an ovum by a sperm
Sperm (: sperm or sperms) is the male reproductive Cell (biology), cell, or gamete, in anisogamous forms of sexual reproduction (forms in which there is a larger, female reproductive cell and a smaller, male one). Animals produce motile sperm ...
. In addition, almost 10% of DNA methylation
DNA methylation is a biological process by which methyl groups are added to the DNA molecule. Methylation can change the activity of a DNA segment without changing the sequence. When located in a gene promoter (genetics), promoter, DNA methylati ...
s in neuron
A neuron (American English), neurone (British English), or nerve cell, is an membrane potential#Cell excitability, excitable cell (biology), cell that fires electric signals called action potentials across a neural network (biology), neural net ...
s of the hippocampus can be rapidly altered during formation of a strong fear memory.
After fertilization in mammals, DNA methylation
DNA methylation is a biological process by which methyl groups are added to the DNA molecule. Methylation can change the activity of a DNA segment without changing the sequence. When located in a gene promoter (genetics), promoter, DNA methylati ...
patterns are largely erased and then re-established during early embryonic development. Almost all of the methylations from the parents are erased, first during early embryogenesis
An embryo ( ) is the initial stage of development for a multicellular organism. In organisms that reproduce sexually, embryonic development is the part of the life cycle that begins just after fertilization of the female egg cell by the male ...
, and again in gametogenesis, with demethylation and remethylation occurring each time. Demethylation during early embryogenesis occurs in the preimplantation period. After a sperm fertilizes an ovum to form a zygote
A zygote (; , ) is a eukaryote, eukaryotic cell (biology), cell formed by a fertilization event between two gametes.
The zygote's genome is a combination of the DNA in each gamete, and contains all of the genetic information of a new individ ...
, rapid DNA demethylation
For molecular biology in mammals, DNA demethylation causes replacement of 5-methylcytosine (5mC) in a DNA sequence by cytosine (C) (see figure of 5mC and C). DNA demethylation can occur by an active process at the site of a 5mC in a DNA sequence ...
of the paternal DNA and slower demethylation of the maternal DNA occurs until formation of a morula
In embryology, cleavage is the division of cells in the early development of the embryo, following fertilization. The zygotes of many species undergo rapid cell cycles with no significant overall growth, producing a cluster of cells the same siz ...
, which has almost no methylation. After the blastocyst
The blastocyst is a structure formed in the early embryonic development of mammals. It possesses an inner cell mass (ICM) also known as the ''embryoblast'' which subsequently forms the embryo, and an outer layer of trophoblast cells called the ...
is formed, methylation can begin, and with formation of the epiblast a wave of methylation then takes place until the implantation stage of the embryo. Another period of rapid and almost complete demethylation occurs during gametogenesis within the primordial germ cell
A germ cell is any cell that gives rise to the gametes of an organism that reproduces sexually. In many animals, the germ cells originate in the primitive streak and migrate via the gut of an embryo to the developing gonads. There, they unde ...
s (PGCs). Other than the PGCs, in the post-implantation stage, methylation patterns in somatic cells are stage- and tissue-specific with changes that presumably define each individual cell type and last stably over a long time.
Embryonic development
 The mouse
The mouse sperm
Sperm (: sperm or sperms) is the male reproductive Cell (biology), cell, or gamete, in anisogamous forms of sexual reproduction (forms in which there is a larger, female reproductive cell and a smaller, male one). Animals produce motile sperm ...
genome
A genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as ...
is 80–90% methylated at its CpG sites in DNA, amounting to about 20 million methylated sites. After fertilization
Fertilisation or fertilization (see American and British English spelling differences#-ise, -ize (-isation, -ization), spelling differences), also known as generative fertilisation, syngamy and impregnation, is the fusion of gametes to give ...
, the paternal chromosome is almost completely demethylated in six hours by an active process, before DNA replication (blue line in Figure). In the mature oocyte
An oocyte (, oöcyte, or ovocyte) is a female gametocyte or germ cell involved in reproduction. In other words, it is an immature ovum, or egg cell. An oocyte is produced in a female fetus in the ovary during female gametogenesis. The female ger ...
, about 40% of its CpG sites are methylated. Demethylation of the maternal chromosome largely takes place by blockage of the methylating enzyme
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different mol ...
s from acting on maternal-origin DNA and by dilution of the methylated maternal DNA during replication (red line in Figure). The morula
In embryology, cleavage is the division of cells in the early development of the embryo, following fertilization. The zygotes of many species undergo rapid cell cycles with no significant overall growth, producing a cluster of cells the same siz ...
(at the 16 cell stage), has only a small amount of DNA methylation
DNA methylation is a biological process by which methyl groups are added to the DNA molecule. Methylation can change the activity of a DNA segment without changing the sequence. When located in a gene promoter (genetics), promoter, DNA methylati ...
(black line in Figure). Methylation begins to increase at 3.5 days after fertilization in the blastocyst
The blastocyst is a structure formed in the early embryonic development of mammals. It possesses an inner cell mass (ICM) also known as the ''embryoblast'' which subsequently forms the embryo, and an outer layer of trophoblast cells called the ...
, and a large wave of methylation then occurs on days 4.5 to 5.5 in the epiblast, going from 12% to 62% methylation, and reaching maximum level after implantation in the uterus. By day seven after fertilization, the newly formed primordial germ cells (PGC) in the implanted embryo
An embryo ( ) is the initial stage of development for a multicellular organism. In organisms that reproduce sexually, embryonic development is the part of the life cycle that begins just after fertilization of the female egg cell by the male sp ...
segregate from the remaining somatic cell
In cellular biology, a somatic cell (), or vegetal cell, is any biological cell forming the body of a multicellular organism other than a gamete, germ cell, gametocyte or undifferentiated stem cell. Somatic cells compose the body of an organism ...
s. At this point the PGCs have about the same level of methylation as the somatic cells.
The newly formed primordial germ cells (PGC) in the implanted embryo devolve from the somatic cells. At this point the PGCs have high levels of methylation. These cells migrate from the epiblast toward the gonadal ridge. Now the cells are rapidly proliferating and beginning demethylation in two waves. In the first wave, demethylation is by replicative dilution, but in the second wave demethylation is by an active process. The second wave leads to demethylation of specific loci. At this point the PGC genomes display the lowest levels of DNA methylation of any cells in the entire life cycle t embryonic day 13.5 (E13.5), see the second figure in this section
 After fertilization some cells of the newly formed embryo migrate to the germinal ridge and will eventually become the
After fertilization some cells of the newly formed embryo migrate to the germinal ridge and will eventually become the germ cell
A germ cell is any cell that gives rise to the gametes of an organism that reproduces sexually. In many animals, the germ cells originate in the primitive streak and migrate via the gut of an embryo to the developing gonads. There, they unde ...
s (sperm and oocytes) of the next generation. Due to the phenomenon of genomic imprinting, maternal and paternal genomes are differentially marked and must be properly reprogrammed every time they pass through the germline. Therefore, during the process of gametogenesis the primordial germ cells must have their original biparental DNA methylation
DNA methylation is a biological process by which methyl groups are added to the DNA molecule. Methylation can change the activity of a DNA segment without changing the sequence. When located in a gene promoter (genetics), promoter, DNA methylati ...
patterns erased and re-established based on the sex of the transmitting parent.
After fertilization, the paternal and maternal genomes are demethylated in order to erase their epigenetic signatures and acquire totipotency
Cell potency is a cell's ability to differentiate into other cell types.
The more cell types a cell can differentiate into, the greater its potency. Potency is also described as the gene activation potential within a cell, which like a continuum ...
. There is asymmetry at this point: the male pronucleus undergoes a quick and active demethylation. Meanwhile the female pronucleus is demethylated passively during consecutive cell divisions. The process of DNA demethylation
For molecular biology in mammals, DNA demethylation causes replacement of 5-methylcytosine (5mC) in a DNA sequence by cytosine (C) (see figure of 5mC and C). DNA demethylation can occur by an active process at the site of a 5mC in a DNA sequence ...
involves base excision repair and likely other DNA-repair-based mechanisms. Despite the global nature of this process, there are certain sequences that avoid it, such as differentially methylated regions (DMRS) associated with imprinted genes, retrotransposons
Retrotransposons (also called Class I transposable elements) are transposable element, mobile elements which move in the host genome by converting their transcribed RNA into DNA through reverse transcription. Thus, they differ from Class II trans ...
and centromeric heterochromatin. Remethylation is needed again to differentiate the embryo into a complete organism.
''In vitro'' manipulation of pre-implantation embryos has been shown to disrupt methylation patterns at imprinted loci and plays a crucial role in cloned animals.
Learning and memory

Learning
Learning is the process of acquiring new understanding, knowledge, behaviors, skills, value (personal and cultural), values, Attitude (psychology), attitudes, and preferences. The ability to learn is possessed by humans, non-human animals, and ...
and memory have levels of permanence, differing from other mental processes such as thought, language, and consciousness, which are temporary in nature. Learning and memory can be either accumulated slowly (multiplication tables) or rapidly (touching a hot stove), but once attained, can be recalled into conscious use for a long time. Rats subjected to one instance of contextual fear conditioning create an especially strong long-term memory. At 24 h after training, 9.17% of the genes in the rat genomes of hippocampus
The hippocampus (: hippocampi; via Latin from Ancient Greek, Greek , 'seahorse'), also hippocampus proper, is a major component of the brain of humans and many other vertebrates. In the human brain the hippocampus, the dentate gyrus, and the ...
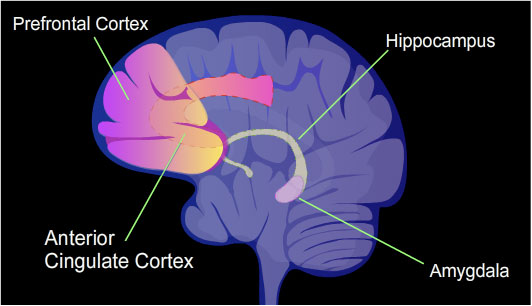
neurons were found to be differentially methylated. This included more than 2,000 differentially methylated genes at 24 hours after training, with over 500 genes being demethylated. The hippocampus region of the brain is where contextual fear memories are first stored (see figure of the brain, this section), but this storage is transient and does not remain in the hippocampus. In rats contextual fear conditioning is abolished when the hippocampus is subjected to hippocampectomy just 1 day after conditioning, but rats retain a considerable amount of contextual fear when a long delay (28 days) is imposed between the time of conditioning and the time of hippocampectomy.
Molecular stages
Three molecular stages are required for reprogramming the DNA methylome. Stage 1: Recruitment. The enzymes needed for reprogramming are recruited to genome sites that require demethylation or methylation. Stage 2: Implementation. The initial enzymatic reactions take place. In the case of methylation, this is a short step that results in the methylation ofcytosine
Cytosine () (symbol C or Cyt) is one of the four nucleotide bases found in DNA and RNA, along with adenine, guanine, and thymine ( uracil in RNA). It is a pyrimidine derivative, with a heterocyclic aromatic ring and two substituents attac ...
to 5-methylcytosine. Stage 3: Base excision DNA repair. The intermediate products of demethylation are catalysed by specific enzymes of the base excision DNA repair pathway that finally restore cystosine in the DNA sequence.
glycosidic bond
A glycosidic bond or glycosidic linkage is a type of ether bond that joins a carbohydrate (sugar) molecule to another group, which may or may not be another carbohydrate.
A glycosidic bond is formed between the hemiacetal or hemiketal group o ...
resulting in an apyrimidinic site (AP site). In an alternative oxidative deamination pathway, 5hmC can be oxidatively deaminated by APOBEC (AID/APOBEC) deaminases to form 5-hydroxymethyluracil (5hmU) or 5mC can be converted to thymine
Thymine () (symbol T or Thy) is one of the four nucleotide bases in the nucleic acid of DNA that are represented by the letters G–C–A–T. The others are adenine, guanine, and cytosine. Thymine is also known as 5-methyluracil, a pyrimidine ...
(Thy). 5hmU can be cleaved by TDG, SMUG1, NEIL1, or MBD4. AP sites and T:G mismatches are then repaired by base excision repair (BER) enzymes to yield cytosine (Cyt).
TET family
The isoforms of the TET enzymes include at least two isoforms of TET1, one ofTET2
Tet methylcytosine dioxygenase 2 (''TET2'') is a human gene. It resides at chromosome 4q24, in a region showing recurrent microdeletions and copy-neutral loss of heterozygosity (CN-LOH) in patients with diverse myeloid malignancies.
Function
' ...
and three isoforms of TET3. The full-length canonical TET1 isoform appears virtually restricted to early embryos, embryonic stem cells and primordial germ cells (PGCs). The dominant TET1 isoform in most somatic tissues, at least in the mouse, arises from alternative promoter usage which gives rise to a short transcript and a truncated protein designated TET1s. The isoforms of TET3 are the full length form TET3FL, a short form splice variant TET3s, and a form that occurs in oocytes and neurons designated TET3o. TET3o is created by alternative promoter use and contains an additional first N-terminal exon
An exon is any part of a gene that will form a part of the final mature RNA produced by that gene after introns have been removed by RNA splicing. The term ''exon'' refers to both the DNA sequence within a gene and to the corresponding sequence ...
coding for 11 amino acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although over 500 amino acids exist in nature, by far the most important are the 22 α-amino acids incorporated into proteins. Only these 22 a ...
s. TET3o only occurs in oocytes and neurons and was not expressed in embryonic stem cells or in any other cell type or adult mouse tissue tested. Whereas TET1 expression can barely be detected in oocytes and zygotes, and TET2 is only moderately expressed, the TET3 variant TET3o shows extremely high levels of expression in oocytes and zygotes, but is nearly absent at the 2-cell stage. It is possible that TET3o, high in neurons, oocytes and zygotes at the one cell stage, is the major TET enzyme utilized when very large scale rapid demethylations occur in these cells.
Recruitment of TET to DNA
The TET enzymes do not specifically bind to 5-methylcytosine except when recruited. Without recruitment or targeting, TET1 predominantly binds to high CG promoters and CpG islands (CGIs) genome-wide by its CXXC domain that can recognize un-methylated CGIs. TET2 does not have an affinity for 5-methylcytosine in DNA. The CXXC domain of the full-length TET3, which is the predominant form expressed in neurons, binds most strongly to CpGs where the C was converted to 5-carboxycytosine (5caC). However, it also binds to un-methylated CpGs.OGG1
Oxoguanine glycosylase (OGG1) catalyses the first step in base excision repair of the oxidatively damaged base 8-OHdG. OGG1 finds 8-OHdG by sliding along the linear DNA at 1,000 base pairs of DNA in 0.1 seconds. OGG1 very rapidly finds 8-OHdG. OGG1 proteins bind to oxidatively damaged DNA with a half maximum time of about 6 seconds. When OGG1 finds 8-OHdG it changes conformation and complexes with 8-OHdG in the binding pocket of OGG1. OGG1 does not immediately act to remove the 8-OHdG. Half maximum removal of 8-OHdG takes about 30 minutes in HeLa cells ''in vitro'', or about 11 minutes in the livers of irradiated mice. DNA oxidation byreactive oxygen species
In chemistry and biology, reactive oxygen species (ROS) are highly Reactivity (chemistry), reactive chemicals formed from diatomic oxygen (), water, and hydrogen peroxide. Some prominent ROS are hydroperoxide (H2O2), superoxide (O2−), hydroxyl ...
preferentially occurs at a guanine
Guanine () (symbol G or Gua) is one of the four main nucleotide bases found in the nucleic acids DNA and RNA, the others being adenine, cytosine, and thymine ( uracil in RNA). In DNA, guanine is paired with cytosine. The guanine nucleoside ...
in a methylated CpG site, because of a lowered ionization potential
In physics and chemistry, ionization energy (IE) is the minimum energy required to remove the most loosely bound electron of an isolated gaseous atom, positive ion, or molecule. The first ionization energy is quantitatively expressed as
:X(g) ...
of guanine bases adjacent to 5-methylcytosine. TET1 binds (is recruited to) the OGG1 bound to 8-OHdG (see figure). This likely allows TET1 to demethylate an adjacent methylated cytosine. When human mammary epithelial cell
Epithelium or epithelial tissue is a thin, continuous, protective layer of Cell (biology), cells with little extracellular matrix. An example is the epidermis, the outermost layer of the skin. Epithelial (Mesothelium, mesothelial) tissues line ...
s (MCF-10A) were treated with H2O2, 8-OHdG increased in DNA by 3.5-fold and this caused large scale demethylation of 5-methylcytosine to about 20% of its initial level in DNA.
EGR1
The gene ''early growth response protein 1'' ('' EGR1'') is an immediate early gene (IEG). The defining characteristic of IEGs is the rapid and transient up-regulation—within minutes—of their mRNA levels independent of protein synthesis. EGR1 can rapidly be induced by neuronal activity. In adulthood, EGR1 is expressed widely throughout the brain, maintaining baseline expression levels in several key areas of the brain including the medial prefrontal cortex,striatum
The striatum (: striata) or corpus striatum is a cluster of interconnected nuclei that make up the largest structure of the subcortical basal ganglia. The striatum is a critical component of the motor and reward systems; receives glutamat ...
, hippocampus and amygdala
The amygdala (; : amygdalae or amygdalas; also '; Latin from Greek language, Greek, , ', 'almond', 'tonsil') is a paired nucleus (neuroanatomy), nuclear complex present in the Cerebral hemisphere, cerebral hemispheres of vertebrates. It is c ...
. This expression is linked to control of cognition, emotional response, social behavior and sensitivity to reward. EGR1 binds to DNA at sites with the motifs 5′-GCGTGGGCG-3′ and 5'-GCGGGGGCGG-3′ and these motifs occur primarily in promoter regions of genes. The short isoform TET1s is expressed in the brain. EGR1 and TET1s form a complex mediated by the C-terminal
The C-terminus (also known as the carboxyl-terminus, carboxy-terminus, C-terminal tail, carboxy tail, C-terminal end, or COOH-terminus) is the end of an amino acid chain (protein or polypeptide), terminated by a free carboxyl group (-COOH). When t ...
regions of both proteins, independently of association with DNA. EGR1 recruits TET1s to genomic regions flanking EGR1 binding sites. In the presence of EGR1, TET1s is capable of locus-specific demethylation and activation of the expression of downstream genes regulated by EGR1.
History
The first person to successfully demonstrate reprogramming was John Gurdon, who in 1962 demonstrated that differentiated somatic cells could be reprogrammed back into an embryonic state when he managed to obtain swimming tadpoles following the transfer of differentiated intestinal epithelial cells into enucleated frog eggs. For this achievement he received the 2012Nobel Prize in Medicine
The Nobel Prize in Physiology or Medicine () is awarded yearly by the Nobel Assembly at the Karolinska Institute, Nobel Assembly at the Karolinska Institute for outstanding discoveries in physiology or medicine. The Nobel Prize is not a single ...
alongside Shinya Yamanaka. Yamanaka was the first to demonstrate (in 2006) that this somatic cell nuclear transfer or oocyte-based reprogramming process (see below), that Gurdon discovered, could be recapitulated (in mice) by defined factors ( Oct4, Sox2, Klf4, and c-Myc) to generate induced pluripotent stem cell
Induced pluripotent stem cells (also known as iPS cells or iPSCs) are a type of pluripotent stem cell that can be generated directly from a somatic cell. The iPSC technology was pioneered by Shinya Yamanaka and Kazutoshi Takahashi in Kyoto, Jap ...
s (iPSCs). Other combinations of genes have also been used, including LIN25 and Homeobox protein NANOG.
Phases of reprogramming
With the discovery that cell fate could be altered, the question of what progression of events occurs signifies a cell undergoing reprogramming. As the final product of iPSC reprogramming was similar in morphology, proliferation,gene expression
Gene expression is the process (including its Regulation of gene expression, regulation) by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, proteins or non-coding RNA, ...
, pluripotency, and telomerase activity, genetic and morphological markers were used as a way to determine what phase of reprogramming was occurring. Reprogramming is defined into three phase: initiation, maturation, and stabilization.
Initiation
The initiation phase is associated with the downregulation of cell type specific genes and the upregulation of pluripotent genes. As the cells move towards pluripotency, the telomerase activity is reactivated to extend telomeres. The cell morphology can directly affect the reprogramming process as the cell is modifying itself to prepare for the gene expression of pluripotency. The main indicator that the initiation phase has completed is that the first genes associated with pluripotency are expressed. This includes the expression of Oct-4 or Homeobox protein NANOG, while undergoing a mesenchymal–epithelial transition (MET), and the loss ofapoptosis
Apoptosis (from ) is a form of programmed cell death that occurs in multicellular organisms and in some eukaryotic, single-celled microorganisms such as yeast. Biochemistry, Biochemical events lead to characteristic cell changes (Morphology (biol ...
and senescence
Senescence () or biological aging is the gradual deterioration of Function (biology), functional characteristics in living organisms. Whole organism senescence involves an increase in mortality rate, death rates or a decrease in fecundity with ...
.
If the cell is directly reprogrammed from one somatic cell
In cellular biology, a somatic cell (), or vegetal cell, is any biological cell forming the body of a multicellular organism other than a gamete, germ cell, gametocyte or undifferentiated stem cell. Somatic cells compose the body of an organism ...
to another, the genes associated with each cell type begin to be upregulated and downregulated accordingly. This can either occur through direct cell reprogramming or creating an intermediate, such as a iPSC, and differentiating into the desired cell type.
The initiation phase is completed through one of three pathways: nuclear transfer, cell fusion Cell fusion is an important cellular process in which several uninucleate cells (cells with a single nucleus) combine to form a multinucleate cell, known as a syncytium. Cell fusion occurs during differentiation of myoblasts, osteoclasts and ...
, or defined factors (microRNA
Micro ribonucleic acid (microRNA, miRNA, μRNA) are small, single-stranded, non-coding RNA molecules containing 21–23 nucleotides. Found in plants, animals, and even some viruses, miRNAs are involved in RNA silencing and post-transcr ...
, transcription factor
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription (genetics), transcription of genetics, genetic information from DNA to messenger RNA, by binding t ...
, epigenetic markers, and other small molecules).
Somatic cell nuclear transfer
 An
An oocyte
An oocyte (, oöcyte, or ovocyte) is a female gametocyte or germ cell involved in reproduction. In other words, it is an immature ovum, or egg cell. An oocyte is produced in a female fetus in the ovary during female gametogenesis. The female ger ...
can reprogram an adult nucleus into an embryonic state after somatic cell nuclear transfer
In genetics and developmental biology, somatic cell nuclear transfer (SCNT) is a laboratory strategy for creating a viable embryo from a body cell and an egg cell. The technique consists of taking a denucleated oocyte (egg cell) and implanti ...
, so that a new organism can be developed from such cell.
Reprogramming is distinct from development of a somatic epitype, as somatic epitypes can potentially be altered after an organism has left the developmental stage of life. During somatic cell nuclear transfer, the oocyte turns off tissue specific genes in the somatic cell nucleus and turns back on embryonic specific genes. This process has been shown through cloning, as seen through John Gurdon with the tadpoles and Dolly the Sheep. Notably, these events have shown that cell fate is a reversible process.
Cell fusion

Cell fusion Cell fusion is an important cellular process in which several uninucleate cells (cells with a single nucleus) combine to form a multinucleate cell, known as a syncytium. Cell fusion occurs during differentiation of myoblasts, osteoclasts and ...
is used to create a multi nucleated cell called a heterokaryon. The fused cells allow for otherwise silenced genes to become reactivated and expressive. As the genes are reactivated, the cells can re-differentiate. There are instances where transcriptional factors, such as the Yamanaka factors, are still needed to aid in heterokaryon cell reprogramming.
Defined factors
 Unlike nuclear transfer and cell fusion, defined factors do not require a full genome, only reprogramming factors. These reprogramming factors include
Unlike nuclear transfer and cell fusion, defined factors do not require a full genome, only reprogramming factors. These reprogramming factors include microRNA
Micro ribonucleic acid (microRNA, miRNA, μRNA) are small, single-stranded, non-coding RNA molecules containing 21–23 nucleotides. Found in plants, animals, and even some viruses, miRNAs are involved in RNA silencing and post-transcr ...
, transcription factor
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription (genetics), transcription of genetics, genetic information from DNA to messenger RNA, by binding t ...
, epigenetic markers, and other small molecules. The original transcription factors, that lead to iPSC development, discovered by Yamanaka include Oct4, Sox2, Klf4, and c-Myc (OSKM factors). Although the OSKM factors have been shown to induce and aid in pluripotency, other transcription factors such as Homeobox protein NANOG, LIN25, TRA-1-60, and C/EBPα aid in the efficiency of reprogramming. The use of microRNA
Micro ribonucleic acid (microRNA, miRNA, μRNA) are small, single-stranded, non-coding RNA molecules containing 21–23 nucleotides. Found in plants, animals, and even some viruses, miRNAs are involved in RNA silencing and post-transcr ...
and other small molecule-driven processes has been utilized as a means of increasing the efficiency of the differentiation from somatic cells to pluripotency.
Maturation
The maturation phase begins at the end of the initiation phase, when the first pluripotent genes are expressed. The cell is preparing itself to be independent from the defined factors, that started the reprogramming process. The first genes to be detected in iPSCs are Oct4, Homeobox protein NANOG, and Esrrb, followed later by Sox2. In the later stages of maturation,transgene
A transgene is a gene that has been transferred naturally, or by any of a number of genetic engineering techniques, from one organism to another. The introduction of a transgene, in a process known as transgenesis, has the potential to change the ...
silencing marks the start of the cell becoming independent from the induced transcription factor
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription (genetics), transcription of genetics, genetic information from DNA to messenger RNA, by binding t ...
. Once the cell is independent, the maturation phase ends and the stabilization phase begins.
As reprogramming efficiency has proven to be a variable and low efficiency process, not all the cells complete the maturation phase and achieve pluripotency. Some cells that undergo reprogramming still remain under apoptosis
Apoptosis (from ) is a form of programmed cell death that occurs in multicellular organisms and in some eukaryotic, single-celled microorganisms such as yeast. Biochemistry, Biochemical events lead to characteristic cell changes (Morphology (biol ...
at the beginning of the maturation stage from oxidative stress
Oxidative stress reflects an imbalance between the systemic manifestation of reactive oxygen species and a biological system's ability to readily detoxify the reactive intermediates or to repair the resulting damage. Disturbances in the normal ...
brought on by the stresses of gene expression change. The use of microRNA
Micro ribonucleic acid (microRNA, miRNA, μRNA) are small, single-stranded, non-coding RNA molecules containing 21–23 nucleotides. Found in plants, animals, and even some viruses, miRNAs are involved in RNA silencing and post-transcr ...
, proteins, and different combinations of the OSKM factors have started to lead towards a higher efficiency rate of reprogramming.
Stabilization
The stabilization phase refers to the processes in the cell that occur after the cell reaches pluripotency. One genetic marker is the expression of Sox2 andX chromosome
The X chromosome is one of the two sex chromosomes in many organisms, including mammals, and is found in both males and females. It is a part of the XY sex-determination system and XO sex-determination system. The X chromosome was named for its u ...
reactivation, while epigenetic changes include the telomerase extending the telomeres and loss of the cell’s epigenetic memory. The epigenetic memory of a cell is reset by the changes in DNA methylation, using activation-induced cytidine deaminase (AID), TET enzymes (TET), and DNA methyltransferase
In biochemistry, the DNA methyltransferase (DNA MTase, DNMT) family of enzymes catalyze the transfer of a methyl group to DNA. DNA methylation serves a wide variety of biological functions. All the known DNA methyltransferases use S-adenosyl ...
(DMNTs), starting in the maturation phase and into the stabilization stage. Once the epigenetic memory of the cell is lost, the possibility of differentiation into the three germ layers is achieved. This is considered a fully reprogrammed cell as it can be passaged without reverting to its original somatic cell type.In cell culture systems
Reprogramming can also be induced artificially through the introduction of exogenous factors, usuallytranscription factor
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription (genetics), transcription of genetics, genetic information from DNA to messenger RNA, by binding t ...
s. In this context, it often refers to the creation of induced pluripotent stem cell
Induced pluripotent stem cells (also known as iPS cells or iPSCs) are a type of pluripotent stem cell that can be generated directly from a somatic cell. The iPSC technology was pioneered by Shinya Yamanaka and Kazutoshi Takahashi in Kyoto, Jap ...
s from mature cells such as adult fibroblast
A fibroblast is a type of cell (biology), biological cell typically with a spindle shape that synthesizes the extracellular matrix and collagen, produces the structural framework (Stroma (tissue), stroma) for animal Tissue (biology), tissues, and ...
s. This allows the production of stem cell
In multicellular organisms, stem cells are undifferentiated or partially differentiated cells that can change into various types of cells and proliferate indefinitely to produce more of the same stem cell. They are the earliest type of cell ...
s for biomedical research
Medical research (or biomedical research), also known as health research, refers to the process of using scientific methods with the aim to produce knowledge about human diseases, the prevention and treatment of illness, and the promotion of ...
, such as research into stem cell therapies, without the use of embryos. It is carried out by the transfection of stem-cell associated genes into mature cells using viral vector
A viral vector is a modified virus designed to gene delivery, deliver genetic material into cell (biology), cells. This process can be performed inside an organism or in cell culture. Viral vectors have widespread applications in basic research, ...
s such as retrovirus
A retrovirus is a type of virus that inserts a DNA copy of its RNA genome into the DNA of a host cell that it invades, thus changing the genome of that cell. After invading a host cell's cytoplasm, the virus uses its own reverse transcriptase e ...
es.
Transcription factors
One of the first transacting factors discovered to change a cell was found in a myoblast when thecomplementary DNA
In genetics, complementary DNA (cDNA) is DNA that was reverse transcribed (via reverse transcriptase) from an RNA (e.g., messenger RNA or microRNA). cDNA exists in both single-stranded and double-stranded forms and in both natural and engin ...
(cDNA) coding for MyoD
MyoD, also known as myoblast determination protein 1, is a protein in animals that plays a major role in regulating muscle differentiation. MyoD, which was discovered in the laboratory of Harold M. Weintraub, belongs to a family of proteins kn ...
was expressed and converted a fibroblast
A fibroblast is a type of cell (biology), biological cell typically with a spindle shape that synthesizes the extracellular matrix and collagen, produces the structural framework (Stroma (tissue), stroma) for animal Tissue (biology), tissues, and ...
to a myoblast. Another transacting factor that directly transformed a lymphoid cell into a myeloid cell
Myeloid tissue, in the bone marrow sense of the word ''wikt:myeloid#Adjective, myeloid'' (''wikt:myelo-#Prefix, myelo-'' + ''wikt:-oid#Suffix, -oid''), is tissue (biology), tissue of bone marrow, of bone marrow cell lineage, or resembling bon ...
was C/EBPα. MyoD and C/EBPα are examples of a small number of single factors that can transform cells. More often, a combination of transcription factors work in conjunction to reprogram a cell.
OSKM
The OSKM factors ( Oct4, Sox2, Klf4, and c-Myc) were initially discovered by Yamanaka in 2006, by the induction of a mouse fibroblast into aninduced pluripotent stem cell
Induced pluripotent stem cells (also known as iPS cells or iPSCs) are a type of pluripotent stem cell that can be generated directly from a somatic cell. The iPSC technology was pioneered by Shinya Yamanaka and Kazutoshi Takahashi in Kyoto, Jap ...
(iPSCs). Within the following year, these factors were used to induce human fibroblasts into iPSCs.
Oct4 is part of the core regulatory genes needed for pluripotency, as it is seen in both embryonic stem cells and tumors. The use of Oct4 even in small increases allows for the start differentiation into pluripotency. Oct4 works in conjecture with Sox2 for the expression of FGF4 which could aid in differentiation.
Sox2 is a gene used in maintaining pluripotency in stem cells. Oct4 and Sox2 work together to regulate hundreds of genes utilized in pluripotency. However, Sox2 is not the only possible Sox family member to participate in gene regulation with Oct4 – Sox4, Sox11, and Sox15 also participate, as the Sox protein is redundant throughout the stem cell genome
A genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as ...
.
Klf4 is a transcription factor used in proliferation, differentiation, apoptosis
Apoptosis (from ) is a form of programmed cell death that occurs in multicellular organisms and in some eukaryotic, single-celled microorganisms such as yeast. Biochemistry, Biochemical events lead to characteristic cell changes (Morphology (biol ...
, and somatic cell
In cellular biology, a somatic cell (), or vegetal cell, is any biological cell forming the body of a multicellular organism other than a gamete, germ cell, gametocyte or undifferentiated stem cell. Somatic cells compose the body of an organism ...
reprogramming. When being utilized in cellular reprogramming, Klf4 prevents cell division of damaged cells using its apoptotic ability, and aids in histone acetyltransferase
Histone acetyltransferases (HATs) are enzymes that acetylation, acetylate conserved lysine amino acids on histone proteins by transferring an acetyl group from acetyl-CoA to form ε-N-acetyllysine, ε-''N''-acetyllysine. DNA is wrapped around his ...
activity.
c-Myc is also known as an oncogene, and in certain conditions can become cancer causing. In cellular reprogramming, c-Myc is used for cell cycle
The cell cycle, or cell-division cycle, is the sequential series of events that take place in a cell (biology), cell that causes it to divide into two daughter cells. These events include the growth of the cell, duplication of its DNA (DNA re ...
progression, apoptosis
Apoptosis (from ) is a form of programmed cell death that occurs in multicellular organisms and in some eukaryotic, single-celled microorganisms such as yeast. Biochemistry, Biochemical events lead to characteristic cell changes (Morphology (biol ...
, and cellular transformation for further differentiation.
NANOG
Homeobox protein NANOG (NANOG) is a transcription factor used to aid in the efficiency of generating iPSCs by maintaining pluripotency and suppressing cell determination factors. NANOG works by promotingchromatin
Chromatin is a complex of DNA and protein found in eukaryote, eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important r ...
accessibility through repression of histone
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei and in most Archaeal phyla. They act as spools around which DNA winds to create structural units called nucleosomes ...
markers, such as H3K27me3
H3K27me3 is an epigenetic modification to the DNA packaging protein histone H3. It is a mark that indicates the tri-methylation of lysine 27 on histone H3 protein.
This tri-methylation is associated with the Downregulation and upregulation, down ...
. NANOG aids recruitment of Oct4, Sox2, and Esrrb used in transcription, while also recruiting Brahma-related gene-1 (BRG1) for chromatin
Chromatin is a complex of DNA and protein found in eukaryote, eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important r ...
accessibility.
C/EBPα
CEBPA is a commonly used factor when reprogramming cells into not only iPSCs, but also other cells. C/EBPα has shown itself to be a single transacting factor during direct reprogramming of a lymphoid cell into a myeloid cell. C/EBPα is considered a 'path breaker' to aid in preparing the cell for intake of the OSKM factors and specific transcription events. C/EBPα has also been shown to increase the efficiency of the reprogramming events.Variability
The properties of cells obtained after reprogramming can vary significantly, in particular among iPSCs. Factors leading to variation in the performance of reprogramming and functional features of end products include genetic background, tissue source, reprogramming factor stoichiometry and stressors related to cell culture.See also
* Induced stem cells *Epigenome editing
Epigenome editing or epigenome engineering is a type of genetic engineering in which the epigenome is modified at specific sites using engineered molecules targeted to those sites (as opposed to whole-genome modifications). Whereas gene editing inv ...
References
{{reflist DNA Epigenetics Induced stem cells