RNA-induced silencing complex on:
[Wikipedia]
[Google]
[Amazon]
The RNA-induced silencing complex, or RISC, is a
 Hannon and his colleagues attempted to identify the RNAi mechanisms involved in
Hannon and his colleagues attempted to identify the RNAi mechanisms involved in


 Major proteins of RISC, Ago2, SND1, and AEG-1, act as crucial contributors to the gene silencing function of the complex.
RISC uses the guide strand of miRNA or siRNA to target complementary 3'-untranslated regions (3'UTR) of mRNA transcripts via
Major proteins of RISC, Ago2, SND1, and AEG-1, act as crucial contributors to the gene silencing function of the complex.
RISC uses the guide strand of miRNA or siRNA to target complementary 3'-untranslated regions (3'UTR) of mRNA transcripts via
 Regardless, it is apparent that Argonaute proteins are present and are essential for function. Furthermore, there are insights into some of the key proteins (in addition to Argonaute) within the complex, which allow RISC to carry out its function.
Regardless, it is apparent that Argonaute proteins are present and are essential for function. Furthermore, there are insights into some of the key proteins (in addition to Argonaute) within the complex, which allow RISC to carry out its function.
 It is as yet unclear how the activated RISC complex locates the mRNA targets in the cell, though it has been shown that the process can occur in situations outside of ongoing protein translation from mRNA.
Endogenously expressed miRNA in metazoans is usually not perfectly complementary to a large number of genes and thus, they modulate expression via translational repression. However, in
It is as yet unclear how the activated RISC complex locates the mRNA targets in the cell, though it has been shown that the process can occur in situations outside of ongoing protein translation from mRNA.
Endogenously expressed miRNA in metazoans is usually not perfectly complementary to a large number of genes and thus, they modulate expression via translational repression. However, in
multiprotein complex
A protein complex or multiprotein complex is a group of two or more associated polypeptide chains. Protein complexes are distinct from multienzyme complexes, in which multiple catalytic domains are found in a single polypeptide chain.
Protein ...
, specifically a ribonucleoprotein, which functions in gene silencing
Gene silencing is the regulation of gene expression in a cell to prevent the expression of a certain gene. Gene silencing can occur during either transcription or translation and is often used in research. In particular, methods used to silence ge ...
via a variety of pathways at the transcriptional and translational levels. Using single-stranded RNA (ssRNA) fragments, such as microRNA
MicroRNA (miRNA) are small, single-stranded, non-coding RNA molecules containing 21 to 23 nucleotides. Found in plants, animals and some viruses, miRNAs are involved in RNA silencing and post-transcriptional regulation of gene expression. mi ...
(miRNA), or double-stranded small interfering RNA
Small interfering RNA (siRNA), sometimes known as short interfering RNA or silencing RNA, is a class of double-stranded RNA at first non-coding RNA molecules, typically 20-24 (normally 21) base pairs in length, similar to MicroRNA, miRNA, and op ...
(siRNA), the complex functions as a key tool in gene regulation. The single strand of RNA acts as a template for RISC to recognize complementary messenger RNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
mRNA is created during the ...
(mRNA) transcript. Once found, one of the proteins in RISC, Argonaute
The Argonaute protein family, first discovered for its evolutionarily conserved stem cell function, plays a central role in RNA silencing processes as essential components of the RNA-induced silencing complex (RISC). RISC is responsible for the ...
, activates and cleaves the mRNA. This process is called RNA interference
RNA interference (RNAi) is a biological process in which RNA molecules are involved in sequence-specific suppression of gene expression by double-stranded RNA, through translational or transcriptional repression. Historically, RNAi was known by ...
(RNAi) and it is found in many eukaryotes
Eukaryotes () are organisms whose cells have a nucleus. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the three domains of life. Bacter ...
; it is a key process in defense against viral infections
A viral disease (or viral infection) occurs when an organism's body is invaded by pathogenic viruses, and infectious virus particles (virions) attach to and enter susceptible cells.
Structural Characteristics
Basic structural characteristics, ...
, as it is triggered by the presence of double-stranded RNA (dsRNA).
Discovery
Thebiochemical
Biochemistry or biological chemistry is the study of chemical processes within and relating to living organisms. A sub-discipline of both chemistry and biology, biochemistry may be divided into three fields: structural biology, enzymology an ...
identification of RISC was conducted by Gregory Hannon and his colleagues at the Cold Spring Harbor Laboratory
Cold Spring Harbor Laboratory (CSHL) is a private, non-profit institution with research programs focusing on cancer, neuroscience, plant biology, genomics, and quantitative biology.
It is one of 68 institutions supported by the Cancer Centers ...
.
This was only a couple of years after the discovery of RNA interference in 1998 by Andrew Fire and Craig Mello, who shared the 2006 Nobel Prize in Physiology or Medicine
The Nobel Prize in Physiology or Medicine is awarded yearly by the Nobel Assembly at the Karolinska Institute for outstanding discoveries in physiology or medicine. The Nobel Prize is not a single prize, but five separate prizes that, accordi ...
.
 Hannon and his colleagues attempted to identify the RNAi mechanisms involved in
Hannon and his colleagues attempted to identify the RNAi mechanisms involved in gene silencing
Gene silencing is the regulation of gene expression in a cell to prevent the expression of a certain gene. Gene silencing can occur during either transcription or translation and is often used in research. In particular, methods used to silence ge ...
, by dsRNAs, in ''Drosophila
''Drosophila'' () is a genus of flies, belonging to the family Drosophilidae, whose members are often called "small fruit flies" or (less frequently) pomace flies, vinegar flies, or wine flies, a reference to the characteristic of many speci ...
'' cells. ''Drosophila'' S2 cells were transfected with a '' lacZ'' expression vector
An expression vector, otherwise known as an expression construct, is usually a plasmid or virus designed for gene expression in cells. The vector is used to introduce a specific gene into a target cell, and can commandeer the cell's mechanism for ...
to quantify gene expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, protein or non-coding RNA, and ultimately affect a phenotype, as the final effect. T ...
with β-galactosidase
β-Galactosidase (EC 3.2.1.23, lactase, beta-gal or β-gal; systematic name β-D-galactoside galactohydrolase), is a glycoside hydrolase enzyme that catalyzes hydrolysis of terminal non-reducing β-D-galactose residues in β-D-galactosides.
β ...
activity. Their results showed co-transfection with ''lacZ'' dsRNA significantly reduced β-galactosidase activity compared to control dsRNA. Therefore, dsRNAs control gene expression via sequence complementarity.
S2 cells were then transfected with ''Drosophila'' cyclin E
Cyclin E is a member of the cyclin family.
Cyclin E binds to G1 phase Cdk2, which is required for the transition from G1 to S phase of the cell cycle that determines initiation of DNA duplication. The Cyclin E/CDK2 complex phosphorylates p27 ...
dsRNA. Cycline E is an essential gene for cell cycle
The cell cycle, or cell-division cycle, is the series of events that take place in a cell that cause it to divide into two daughter cells. These events include the duplication of its DNA (DNA replication) and some of its organelles, and sub ...
progression into the S phase
S phase (Synthesis Phase) is the phase of the cell cycle in which DNA is replicated, occurring between G1 phase and G2 phase. Since accurate duplication of the genome is critical to successful cell division, the processes that occur during ...
. Cyclin E dsRNA arrested the cell cycle at the G1 phase (before the S phase). Therefore, RNAi can target endogenous
Endogenous substances and processes are those that originate from within a living system such as an organism, tissue, or cell.
In contrast, exogenous substances and processes are those that originate from outside of an organism.
For example, ...
genes.
In addition, cyclin E dsRNA only diminished cyclin E RNA — a similar result was also shown using dsRNA corresponding to cyclin A
Cyclin A is a member of the cyclin family, a group of proteins that function in regulating progression through the cell cycle. The stages that a cell passes through that culminate in its division and replication are collectively known as the cel ...
which acts in S, G2 and M phases of the cell cycle. This shows the characteristic hallmark of RNAi: the reduced levels of mRNAs correspond to the levels of dsRNA added.
To test whether their observation of decreased mRNA levels was a result of mRNA being targeted directly (as suggested by data from other systems), ''Drosophila'' S2 cells were transfected with either ''Drosophila'' cyclin E dsRNAs or ''lacZ'' dsRNAs and then incubated with synthetic mRNAs for cyclin E or ''lacZ''.
Cells transfected with cyclin E dsRNAs only showed degradation in cyclin E transcripts — the ''lacZ'' transcripts were stable. Conversely, cells transfected with ''lacZ'' dsRNAs only showed degradation in ''lacZ'' transcripts and not cyclin E transcripts. Their results led Hannon and his colleagues to suggest RNAi degrades target mRNA through a 'sequence-specific nuclease
A nuclease (also archaically known as nucleodepolymerase or polynucleotidase) is an enzyme capable of cleaving the phosphodiester bonds between nucleotides of nucleic acids. Nucleases variously effect single and double stranded breaks in their t ...
activity'. They termed the nuclease enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products ...
RISC. Later Devanand Sarkar and his colleagues Prasanna K. Santhekadur and Byoung Kwon Yoo at the Virginia Commonwealth University elucidated the RISC activity and its molecular mechanism in cancer cells and they identified another new component of the RISC, called AEG-1 7
Function in RNA interference

Incorporation of siRNA/miRNA
The RNase III Dicer is a critical member of RISC that initiates the RNA interference process by producing double-stranded siRNA or single-stranded miRNA. Enzymatic cleavage of dsRNA within the cell produces the short siRNA fragments of 21-23nucleotide
Nucleotides are organic molecules consisting of a nucleoside and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of which are essential biomolecu ...
s in length with a two-nucleotide 3' overhang. Dicer also processes pre-miRNA, which forms a hairpin loop structure to mimic dsRNA, in a similar fashion. dsRNA fragments are loaded into RISC with each strand having a different fate based on the asymmetry rule phenomenon, the selection of one strand as the guide strand over the other based on thermodynamic stability. The newly generated miRNA or siRNA act as single-stranded guide sequences for RISC to target mRNA for degradation.
* The strand with the less thermodynamically stable 5' end
Directionality, in molecular biology and biochemistry, is the end-to-end chemical orientation of a single strand of nucleic acid. In a single strand of DNA or RNA, the chemical convention of naming carbon atoms in the nucleotide pentose-sugar- ...
is selected by the protein Argonaute
The Argonaute protein family, first discovered for its evolutionarily conserved stem cell function, plays a central role in RNA silencing processes as essential components of the RNA-induced silencing complex (RISC). RISC is responsible for the ...
and integrated into RISC. This strand is known as the guide strand and targets mRNA for degradation.
* The other strand, known as the passenger strand, is degraded by RISC.

Gene regulation
Watson-Crick base pairing
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both ...
, allowing it to regulate gene expression of the mRNA transcript in a number of ways.
mRNA degradation
The most understood function of RISC is degradation of target mRNA which reduces the levels of transcript available to be translated by ribosomes. The endonucleolytic cleavage of the mRNA complementary to the RISC's guide strand by Argonaute protein is the key to RNAi initiation. There are two main requirements for mRNA degradation to take place: * a near-perfect complementary match between the guide strand and target mRNA sequence, and, * a catalytically active Argonaute protein, called a 'slicer', to cleave the target mRNA. There are two major pathways of mRNA degradation once cleavage has occurred. Both are initiated through degradation of the mRNA's poly(A) tail, resulting in removal of the mRNA's 5' cap. * 5'-to-3' degradation of the transcript occurs by XRN1 exonuclease incytoplasm
In cell biology, the cytoplasm is all of the material within a eukaryotic cell, enclosed by the cell membrane, except for the cell nucleus. The material inside the nucleus and contained within the nuclear membrane is termed the nucleoplasm. ...
ic bodies called P-bodies
P-bodies, or processing bodies are distinct foci formed by phase separation within the cytoplasm of the eukaryotic cell consisting of many enzymes involved in mRNA turnover. P-bodies are highly conserved structures and have been observed in so ...
.
* 3'-to-5' degradation of the transcript is conducted by the exosome and Ski complex The Ski complex is a multi-protein complex involved in the 3' end degradation of messenger RNAs in yeast.
Structure
The complex consists of three main proteins, the RNA helicase ''Ski2'' and the proteins ''Ski3'' and ''Ski8''. This tetramer con ...
.
Translational repression
RISC can modulate the loading of ribosome and accessory factors intranslation
Translation is the communication of the meaning of a source-language text by means of an equivalent target-language text. The English language draws a terminological distinction (which does not exist in every language) between ''transla ...
to repress expression of the bound mRNA transcript. Translational repression only requires a partial sequence match between the guide strand and target mRNA.
Translation can be regulated at the initiation step by:
*preventing the binding of the eukaryotic translation initiation factor (eIF) to the 5' cap
In molecular biology, the five-prime cap (5′ cap) is a specially altered nucleotide on the 5′ end of some primary transcripts such as precursor messenger RNA. This process, known as mRNA capping, is highly regulated and vital in the creation ...
. It has been noted RISC can deadenylate the 3' poly(A) tail which might contribute to repression via the 5' cap.
* preventing the binding of the 60S ribosomal subunit binding to the mRNA can repress translation.
Translation can be regulated at post-initiation steps by:
* peptide degradation,
*promoting premature termination of translation ribosomes, or,
* slowing elongation.
There is still speculation on whether translational repression via initiation and post-initiation is mutually exclusive.
Heterochromatin formation
Some RISCs are able to directly target thegenome
In the fields of molecular biology and genetics, a genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding ...
by recruiting histone methyltransferase
Histone methyltransferases (HMT) are histone-modifying enzymes (e.g., histone-lysine N-methyltransferases and histone-arginine N-methyltransferases), that catalyze the transfer of one, two, or three methyl groups to lysine and arginine residues ...
s to form heterochromatin
Heterochromatin is a tightly packed form of DNA or '' condensed DNA'', which comes in multiple varieties. These varieties lie on a continue between the two extremes of constitutive heterochromatin and facultative heterochromatin. Both play a rol ...
at the gene locus, silencing the gene. These RISCs take the form of a RNA-induced transcriptional silencing complex (RITS). The best studied example is with the yeast
Yeasts are eukaryotic, single-celled microorganisms classified as members of the fungus kingdom. The first yeast originated hundreds of millions of years ago, and at least 1,500 species are currently recognized. They are estimated to constit ...
RITS.
RITS has been shown to direct heterochromatin formation at centromeres through recognition of centromeric repeats. Through base-pairing of siRNA (guide strand) to target chromatin sequences, histone-modifying enzymes can be recruited.
The mechanism is not well understood; however, RITS degrade nascent mRNA transcripts. It has been suggested this mechanism acts as a 'self-reinforcing feedback loop
Feedback occurs when outputs of a system are routed back as inputs as part of a chain of cause-and-effect that forms a circuit or loop. The system can then be said to ''feed back'' into itself. The notion of cause-and-effect has to be handled c ...
' as the degraded nascent transcripts are used by RNA-dependent RNA polymerase
RNA-dependent RNA polymerase (RdRp) or RNA replicase is an enzyme that catalyzes the replication of RNA from an RNA template. Specifically, it catalyzes synthesis of the RNA strand complementary to a given RNA template. This is in contrast to ...
(RdRp) to generate more siRNAs.
In '' Schizosaccharomyces pombe'' and ''Arabidopsis
''Arabidopsis'' (rockcress) is a genus in the family Brassicaceae. They are small flowering plants related to cabbage and mustard. This genus is of great interest since it contains thale cress (''Arabidopsis thaliana''), one of the model organ ...
'', the processing of dsRNA targets into siRNA by Dicer RNases can initiate a gene silencing pathway by heterochromatin formation. An Argonaute protein known as AGO4
The Argonaute protein family, first discovered for its evolutionarily conserved stem cell function, plays a central role in RNA silencing processes as essential components of the RNA-induced silencing complex (RISC). RISC is responsible for the ...
interacts with the small RNAs that define heterochromatic sequences. A histone methyl transferase (HMT), H3K9
The histone code is a hypothesis that the transcription of genetic information encoded in DNA is in part regulated by chemical modifications (known as ''histone marks'') to histone proteins, primarily on their unstructured ends. Together with sim ...
, methylates histone H3 and recruits chromodomain proteins to the methylation sites. DNA methylation maintains the silencing of genes as the heterochromatin sequences can be established or spread.
DNA elimination
The siRNA generated by RISCs seem to have a role in degrading DNA during somaticmacronucleus
A macronucleus (formerly also meganucleus) is the larger type of nucleus in ciliates. Macronuclei are polyploid and undergo direct division without mitosis. It controls the non-reproductive cell functions, such as metabolism. During conjugati ...
development in protozoa
Protozoa (singular: protozoan or protozoon; alternative plural: protozoans) are a group of single-celled eukaryotes, either free-living or parasitic, that feed on organic matter such as other microorganisms or organic tissues and debris. Histo ...
'' Tetrahymena''. It is similar to the epigenetic control of heterochromatin formation and is implied as a defense against invading genetic elements.
Similar to heterochromatin formation in ''S. pombe'' and ''Arabidopsis'', a ''Tetrahymena'' protein related to the Argonaute family, Twi1p, catalyzes DNA elimination of target sequences known as internal elimination sequences (IESs). Using methyltransferases and chromodomain proteins, IESs are heterochromatized and eliminated from the DNA.
RISC-associated proteins
The complete structure of RISC is still unsolved. Many studies have reported a range of sizes and components for RISC but it is not entirely sure whether this is due to there being a number of RISC complexes or due to the different sources that different studies use. Ago, Argonaute; Dcr, Dicer; Dmp68, ''D. melanogaster'' orthologue of mammalian p68 RNA unwindase; eIF2C1, eukaryotic translation initiation factor 2C1; eIF2C2, eukaryotic translation initiation factor 2C2; Fmr1/Fxr, ''D. melanogaster'' orthologue of the fragile-X mental retardation protein; miRNP, miRNA-protein complex; NR, not reported; Tsn, Tudor-staphylococcal nuclease; Vig, vasa intronic gene. Regardless, it is apparent that Argonaute proteins are present and are essential for function. Furthermore, there are insights into some of the key proteins (in addition to Argonaute) within the complex, which allow RISC to carry out its function.
Regardless, it is apparent that Argonaute proteins are present and are essential for function. Furthermore, there are insights into some of the key proteins (in addition to Argonaute) within the complex, which allow RISC to carry out its function.
Argonaute proteins
Argonaute proteins are a family of proteins found inprokaryotes
A prokaryote () is a single-celled organism that lacks a nucleus and other membrane-bound organelles. The word ''prokaryote'' comes from the Greek πρό (, 'before') and κάρυον (, 'nut' or 'kernel').Campbell, N. "Biology:Concepts & Con ...
and eukaryotes. Their function in prokaryotes is unknown but in eukaryotes they are responsible for RNAi. There are eight family members in human Argonautes of which only Argonaute 2 is exclusively involved in targeted RNA cleavage in RISC.

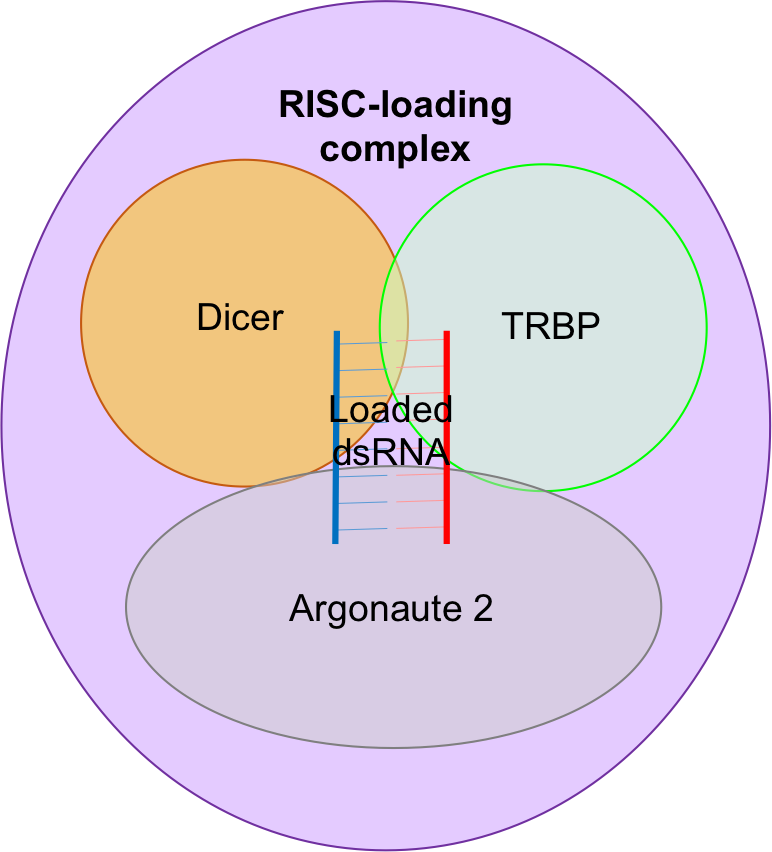
RISC-loading complex
The RISC-loading complex (RLC) is the essential structure required to load dsRNA fragments into RISC in order to target mRNA. The RLC consists of dicer, the transactivating response RNA-binding protein ( TRBP) and Argonaute 2. * Dicer is an RNase IIIendonuclease
Endonucleases are enzymes that cleave the phosphodiester bond within a polynucleotide chain. Some, such as deoxyribonuclease I, cut DNA relatively nonspecifically (without regard to sequence), while many, typically called restriction endonuclease ...
which generates the dsRNA fragments to be loaded that direct RNAi.
* TRBP is a protein with three double-stranded RNA-binding domains.
* Argonaute 2 is an RNase and is the catalytic centre of RISC.
Dicer associates with TRBP and Argonaute 2 to facilitate the transfer of the dsRNA fragments generated by Dicer to Argonaute 2.
More recent research has shown the human RNA helicase A could help facilitate the RLC.
Other proteins
Recently identified members of RISC areSND1
Staphylococcal nuclease domain-containing protein 1 also known as 100 kDa coactivator or Tudor domain-containing protein 11 (TDRD11) is a protein that in humans is encoded by the ''SND1'' gene. SND1 is a main component of RISC complex and plays ...
and MTDH. SND1 and MTDH are oncogenes and regulate various gene expression.
Ago, Argonaute; Dcr, Dicer; Dmp68, ''D. melanogaster'' orthologue of mammalian p68 RNA unwindase; eIF2C1, eukaryotic translation initiation factor 2C1; eIF2C2, eukaryotic translation initiation factor 2C2; Fmr1/Fxr, ''D. melanogaster'' orthologue of the fragile-X mental retardation protein; Tsn, Tudor-staphylococcal nuclease; Vig, vasa intronic gene.
Binding of mRNA
plant
Plants are predominantly photosynthetic eukaryotes of the kingdom Plantae. Historically, the plant kingdom encompassed all living things that were not animals, and included algae and fungi; however, all current definitions of Plantae excl ...
s, the process has a much greater specificity to target mRNA and usually each miRNA only binds to one mRNA. A greater specificity means mRNA degradation is more likely to occur.
See also
*RNA-induced transcriptional silencing
RNA-induced transcriptional silencing (RITS) is a form of RNA interference by which short RNA molecules – such as small interfering RNA (siRNA) – trigger the downregulation of transcription of a particular gene or genomic region. This is us ...
(RITS)
*RNA interference
RNA interference (RNAi) is a biological process in which RNA molecules are involved in sequence-specific suppression of gene expression by double-stranded RNA, through translational or transcriptional repression. Historically, RNAi was known by ...
References
Further reading
* * *External links
* {{Nucleases RNA