Quantitative genetics is the study of
quantitative trait
Complex traits are phenotypes that are controlled by two or more genes and do not follow Mendelian inheritance, Mendel's Law of Dominance. They may have a range of Gene expression, expression which is typically continuous. Both environmental and g ...
s, which are
phenotype
In genetics, the phenotype () is the set of observable characteristics or traits of an organism. The term covers the organism's morphology (physical form and structure), its developmental processes, its biochemical and physiological propert ...
s that vary continuously—such as height or mass—as opposed to phenotypes and gene-products that are
discretely identifiable—such as eye-colour, or the presence of a particular biochemical.
Both of these branches of
genetics
Genetics is the study of genes, genetic variation, and heredity in organisms.Hartl D, Jones E (2005) It is an important branch in biology because heredity is vital to organisms' evolution. Gregor Mendel, a Moravian Augustinians, Augustinian ...
use the frequencies of different
allele
An allele is a variant of the sequence of nucleotides at a particular location, or Locus (genetics), locus, on a DNA molecule.
Alleles can differ at a single position through Single-nucleotide polymorphism, single nucleotide polymorphisms (SNP), ...
s of a
gene
In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protei ...
in breeding populations (gamodemes), and combine them with concepts from simple
Mendelian inheritance
Mendelian inheritance (also known as Mendelism) is a type of biological inheritance following the principles originally proposed by Gregor Mendel in 1865 and 1866, re-discovered in 1900 by Hugo de Vries and Carl Correns, and later popularize ...
to analyze inheritance patterns across generations and descendant lines. While
population genetics
Population genetics is a subfield of genetics that deals with genetic differences within and among populations, and is a part of evolutionary biology. Studies in this branch of biology examine such phenomena as Adaptation (biology), adaptation, s ...
can focus on particular genes and their subsequent metabolic products, quantitative genetics focuses more on the outward phenotypes, and makes only summaries of the underlying genetics.
Due to the continuous distribution of phenotypic values, quantitative genetics must employ many other statistical methods (such as the ''effect size'', the ''mean'' and the ''variance'') to link phenotypes (attributes) to genotypes. Some phenotypes may be analyzed either as discrete categories or as continuous phenotypes, depending on the definition of cut-off points, or on the ''metric'' used to quantify them.
Mendel himself had to discuss this matter in his famous paper,
especially with respect to his peas' attribute ''tall/dwarf'', which actually was derived by adding a cut-off point to "length of stem".
Analysis of
quantitative trait loci
A quantitative trait locus (QTL) is a Locus (genetics), locus (section of DNA) that correlates with variation of a quantitative trait in the phenotype of a Population genetics, population of organisms. QTLs are mapped by identifying which molecula ...
, or QTLs,
is a more recent addition to quantitative genetics, linking it more directly to
molecular genetics
Molecular genetics is a branch of biology that addresses how differences in the structures or expression of DNA molecules manifests as variation among organisms. Molecular genetics often applies an "investigative approach" to determine the st ...
.
Gene effects
In
diploid
Ploidy () is the number of complete sets of chromosomes in a cell, and hence the number of possible alleles for autosomal and pseudoautosomal genes. Here ''sets of chromosomes'' refers to the number of maternal and paternal chromosome copies, ...
organisms, the average
genotypic "value" (locus value) may be defined by the allele "effect" together with a
dominance effect, and also by how genes interact with genes at other loci (
epistasis
Epistasis is a phenomenon in genetics in which the effect of a gene mutation is dependent on the presence or absence of mutations in one or more other genes, respectively termed modifier genes. In other words, the effect of the mutation is depe ...
). The founder of quantitative genetics -
Sir Ronald Fisher - perceived much of this when he proposed the first mathematics of this branch of genetics.

Being a statistician, he defined the gene effects as deviations from a central value—enabling the use of statistical concepts such as mean and variance, which use this idea.
The central value he chose for the gene was the midpoint between the two opposing homo
zygote
A zygote (; , ) is a eukaryote, eukaryotic cell (biology), cell formed by a fertilization event between two gametes.
The zygote's genome is a combination of the DNA in each gamete, and contains all of the genetic information of a new individ ...
s at the one locus. The deviation from there to the "greater" homozygous genotype can be named "''+a''"; and therefore it is "''-a''" from that same midpoint to the "lesser" homozygote genotype. This is the "allele" effect mentioned above. The heterozygote deviation from the same midpoint can be named "''d''", this being the "dominance" effect referred to above. The diagram depicts the idea. However, in reality we measure phenotypes, and the figure also shows how observed phenotypes relate to the gene effects. Formal definitions of these effects recognize this phenotypic focus. Epistasis has been approached statistically as interaction (i.e., inconsistencies),
but ''epigenetics'' suggests a new approach may be needed.
If 0
a was known as "over-dominance".
Allele and genotype frequencies
To obtain means, variances and other statistics, both ''quantities'' and their ''occurrences'' are required. The gene effects (above) provide the framework for ''quantities'': and the ''frequencies'' of the contrasting alleles in the fertilization gamete-pool provide the information on ''occurrences''. 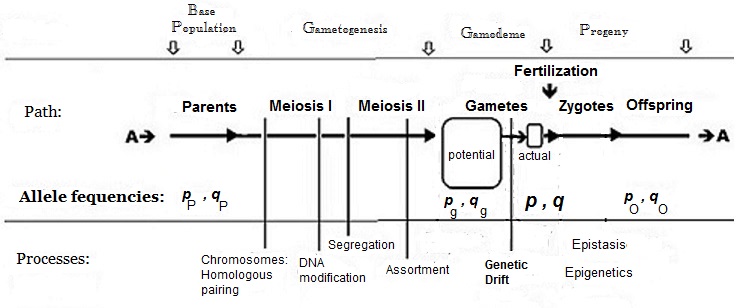 Commonly, the frequency of the allele causing "more" in the phenotype (including dominance) is given the symbol ''p'', while the frequency of the contrasting allele is ''q''. An initial assumption made when establishing the algebra was that the parental population was infinite and random mating, which was made simply to facilitate the derivation. The subsequent mathematical development also implied that the frequency distribution within the effective gamete-pool was uniform: there were no local perturbations where ''p'' and ''q'' varied. Looking at the diagrammatic analysis of sexual reproduction, this is the same as declaring that ''pP'' = ''pg'' = ''p''; and similarly for ''q''.
Commonly, the frequency of the allele causing "more" in the phenotype (including dominance) is given the symbol ''p'', while the frequency of the contrasting allele is ''q''. An initial assumption made when establishing the algebra was that the parental population was infinite and random mating, which was made simply to facilitate the derivation. The subsequent mathematical development also implied that the frequency distribution within the effective gamete-pool was uniform: there were no local perturbations where ''p'' and ''q'' varied. Looking at the diagrammatic analysis of sexual reproduction, this is the same as declaring that ''pP'' = ''pg'' = ''p''; and similarly for ''q''.genetic drift
Genetic drift, also known as random genetic drift, allelic drift or the Wright effect, is the change in the Allele frequency, frequency of an existing gene variant (allele) in a population due to random chance.
Genetic drift may cause gene va ...
'', and is considered subsequently.
While panmixia may not be widely extant, the ''potential'' for it does occur, although it may be only ephemeral because of those local perturbations. It has been shown, for example, that the F2 derived from ''random fertilization of F1 individuals'' (an ''allogamous'' F2), following hybridization, is an ''origin'' of a new ''potentially'' panmictic population.
Random fertilization
Male and female gametes within the actual fertilizing pool are considered usually to have the same frequencies for their corresponding alleles. (Exceptions have been considered.) This means that when ''p'' male gametes carrying the ''A'' allele randomly fertilize ''p'' female gametes carrying that same allele, the resulting zygote has genotype ''AA'', and, under random fertilization, the combination occurs with a frequency of ''p'' x ''p'' (= ''p2''). Similarly, the zygote ''aa'' occurs with a frequency of ''q2''. Heterozygotes (''Aa'') can arise in two ways: when ''p'' male (''A'' allele) randomly fertilize ''q'' female (''a'' allele) gametes, and ''vice versa''. The resulting frequency for the heterozygous zygotes is thus ''2pq''.
Mendel's research cross – a contrast
Mendel's pea experiments were constructed by establishing true-breeding parents with "opposite" phenotypes for each attribute.
Self fertilization – an alternative
Having noticed that the pea is naturally self-pollinated, we cannot continue to use it as an example for illustrating random fertilization properties. Self-fertilization ("selfing") is a major alternative to random fertilization, especially within Plants. Most of the Earth's cereals are naturally self-pollinated (rice, wheat, barley, for example), as well as the pulses. Considering the millions of individuals of each of these on Earth at any time, it is obvious that self-fertilization is at least as significant as random fertilization. Self-fertilization is the most intensive form of ''inbreeding'', which arises whenever there is restricted independence in the genetical origins of gametes. Such reduction in independence arises if parents are already related, and/or from genetic drift or other spatial restrictions on gamete dispersal. Path analysis demonstrates that these are tantamount to the same thing. Being a statistician, he defined the gene effects as deviations from a central value—enabling the use of statistical concepts such as mean and variance, which use this idea. The central value he chose for the gene was the midpoint between the two opposing homo
Being a statistician, he defined the gene effects as deviations from a central value—enabling the use of statistical concepts such as mean and variance, which use this idea. The central value he chose for the gene was the midpoint between the two opposing homo Commonly, the frequency of the allele causing "more" in the phenotype (including dominance) is given the symbol ''p'', while the frequency of the contrasting allele is ''q''. An initial assumption made when establishing the algebra was that the parental population was infinite and random mating, which was made simply to facilitate the derivation. The subsequent mathematical development also implied that the frequency distribution within the effective gamete-pool was uniform: there were no local perturbations where ''p'' and ''q'' varied. Looking at the diagrammatic analysis of sexual reproduction, this is the same as declaring that ''pP'' = ''pg'' = ''p''; and similarly for ''q''. This mating system, dependent upon these assumptions, became known as "panmixia".
Panmixia rarely actually occurs in nature, as gamete distribution may be limited, for example by dispersal restrictions or by behaviour, or by chance sampling (those local perturbations mentioned above). It is well known that there is a huge wastage of gametes in Nature, which is why the diagram depicts a ''potential'' gamete-pool separately to the ''actual'' gamete-pool. Only the latter sets the definitive frequencies for the zygotes: this is the true "gamodeme" ("gamo" refers to the gametes, and "deme" derives from Greek for "population"). But, under Fisher's assumptions, the ''gamodeme'' can be effectively extended back to the ''potential'' gamete-pool, and even back to the parental base-population (the "source" population). The random sampling arising when small "actual" gamete-pools are sampled from a large "potential" gamete-pool is known as ''
Commonly, the frequency of the allele causing "more" in the phenotype (including dominance) is given the symbol ''p'', while the frequency of the contrasting allele is ''q''. An initial assumption made when establishing the algebra was that the parental population was infinite and random mating, which was made simply to facilitate the derivation. The subsequent mathematical development also implied that the frequency distribution within the effective gamete-pool was uniform: there were no local perturbations where ''p'' and ''q'' varied. Looking at the diagrammatic analysis of sexual reproduction, this is the same as declaring that ''pP'' = ''pg'' = ''p''; and similarly for ''q''. This mating system, dependent upon these assumptions, became known as "panmixia".
Panmixia rarely actually occurs in nature, as gamete distribution may be limited, for example by dispersal restrictions or by behaviour, or by chance sampling (those local perturbations mentioned above). It is well known that there is a huge wastage of gametes in Nature, which is why the diagram depicts a ''potential'' gamete-pool separately to the ''actual'' gamete-pool. Only the latter sets the definitive frequencies for the zygotes: this is the true "gamodeme" ("gamo" refers to the gametes, and "deme" derives from Greek for "population"). But, under Fisher's assumptions, the ''gamodeme'' can be effectively extended back to the ''potential'' gamete-pool, and even back to the parental base-population (the "source" population). The random sampling arising when small "actual" gamete-pools are sampled from a large "potential" gamete-pool is known as '' Being a statistician, he defined the gene effects as deviations from a central value—enabling the use of statistical concepts such as mean and variance, which use this idea. The central value he chose for the gene was the midpoint between the two opposing homo
Being a statistician, he defined the gene effects as deviations from a central value—enabling the use of statistical concepts such as mean and variance, which use this idea. The central value he chose for the gene was the midpoint between the two opposing homo Commonly, the frequency of the allele causing "more" in the phenotype (including dominance) is given the symbol ''p'', while the frequency of the contrasting allele is ''q''. An initial assumption made when establishing the algebra was that the parental population was infinite and random mating, which was made simply to facilitate the derivation. The subsequent mathematical development also implied that the frequency distribution within the effective gamete-pool was uniform: there were no local perturbations where ''p'' and ''q'' varied. Looking at the diagrammatic analysis of sexual reproduction, this is the same as declaring that ''pP'' = ''pg'' = ''p''; and similarly for ''q''. This mating system, dependent upon these assumptions, became known as "panmixia".
Panmixia rarely actually occurs in nature, as gamete distribution may be limited, for example by dispersal restrictions or by behaviour, or by chance sampling (those local perturbations mentioned above). It is well known that there is a huge wastage of gametes in Nature, which is why the diagram depicts a ''potential'' gamete-pool separately to the ''actual'' gamete-pool. Only the latter sets the definitive frequencies for the zygotes: this is the true "gamodeme" ("gamo" refers to the gametes, and "deme" derives from Greek for "population"). But, under Fisher's assumptions, the ''gamodeme'' can be effectively extended back to the ''potential'' gamete-pool, and even back to the parental base-population (the "source" population). The random sampling arising when small "actual" gamete-pools are sampled from a large "potential" gamete-pool is known as ''
Commonly, the frequency of the allele causing "more" in the phenotype (including dominance) is given the symbol ''p'', while the frequency of the contrasting allele is ''q''. An initial assumption made when establishing the algebra was that the parental population was infinite and random mating, which was made simply to facilitate the derivation. The subsequent mathematical development also implied that the frequency distribution within the effective gamete-pool was uniform: there were no local perturbations where ''p'' and ''q'' varied. Looking at the diagrammatic analysis of sexual reproduction, this is the same as declaring that ''pP'' = ''pg'' = ''p''; and similarly for ''q''. This mating system, dependent upon these assumptions, became known as "panmixia".
Panmixia rarely actually occurs in nature, as gamete distribution may be limited, for example by dispersal restrictions or by behaviour, or by chance sampling (those local perturbations mentioned above). It is well known that there is a huge wastage of gametes in Nature, which is why the diagram depicts a ''potential'' gamete-pool separately to the ''actual'' gamete-pool. Only the latter sets the definitive frequencies for the zygotes: this is the true "gamodeme" ("gamo" refers to the gametes, and "deme" derives from Greek for "population"). But, under Fisher's assumptions, the ''gamodeme'' can be effectively extended back to the ''potential'' gamete-pool, and even back to the parental base-population (the "source" population). The random sampling arising when small "actual" gamete-pools are sampled from a large "potential" gamete-pool is known as ''