Point Accepted Mutation on:
[Wikipedia]
[Google]
[Amazon]
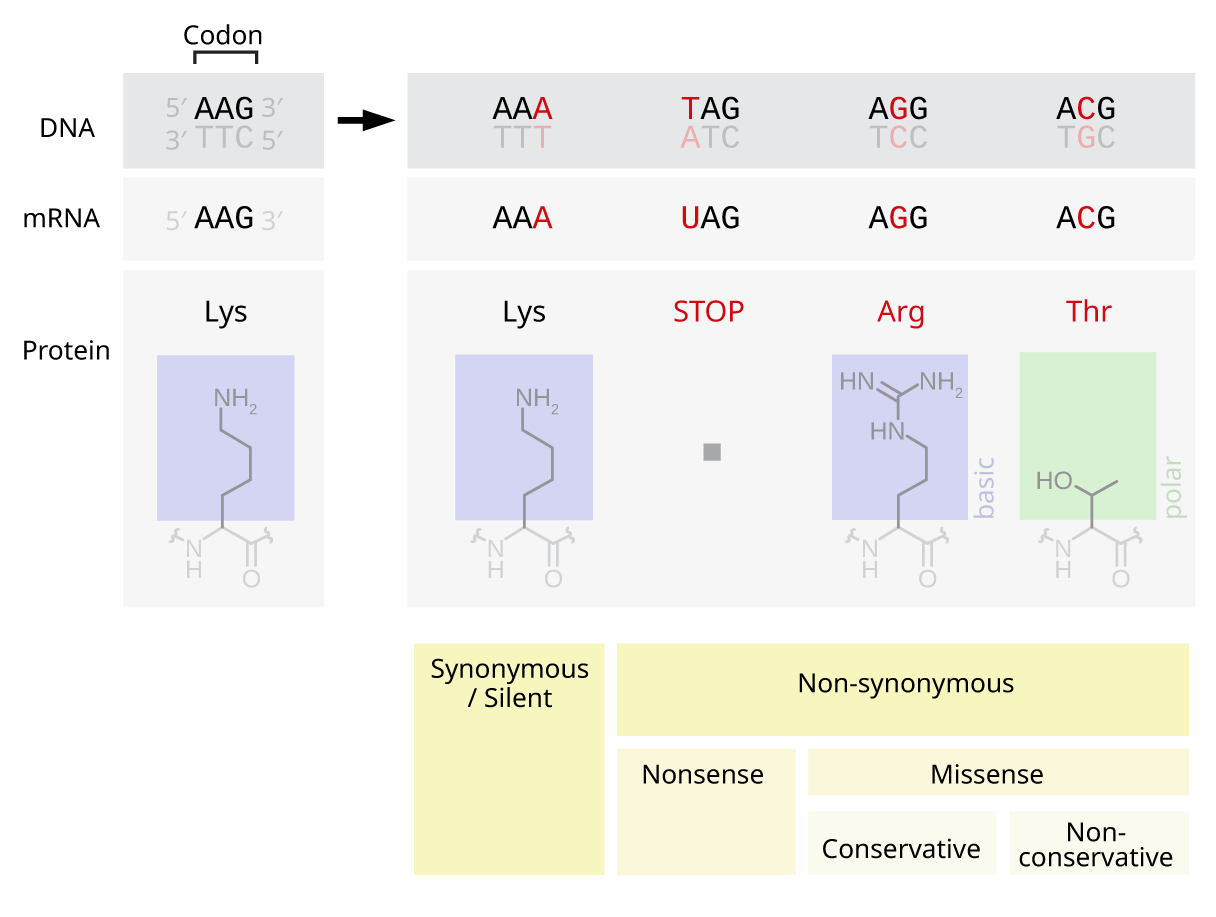 A point accepted mutation — also known as a PAM — is the replacement of a single
A point accepted mutation — also known as a PAM — is the replacement of a single

 A point accepted mutation — also known as a PAM — is the replacement of a single
A point accepted mutation — also known as a PAM — is the replacement of a single amino acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although hundreds of amino acids exist in nature, by far the most important are the alpha-amino acids, which comprise proteins. Only 22 alpha am ...
in the primary structure
Protein primary structure is the linear sequence of amino acids in a peptide or protein. By convention, the primary structure of a protein is reported starting from the amino-terminal (N) end to the carboxyl-terminal (C) end. Protein biosynthes ...
of a protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, respo ...
with another single amino acid, which is accepted by the processes of natural selection
Natural selection is the differential survival and reproduction of individuals due to differences in phenotype. It is a key mechanism of evolution, the change in the heritable traits characteristic of a population over generations. Charle ...
. This definition does not include all point mutations
A point mutation is a genetic mutation where a single nucleotide base is changed, inserted or deleted from a DNA or RNA sequence of an organism's genome. Point mutations have a variety of effects on the downstream protein product—consequence ...
in the DNA of an organism. In particular, silent mutations
Silent mutations are mutations in DNA that do not have an observable effect on the organism's phenotype. They are a specific type of neutral mutation. The phrase ''silent mutation'' is often used interchangeably with the phrase '' synonymous mutat ...
are not point accepted mutations, nor are mutations that are lethal or that are rejected by natural selection in other ways.
A PAM matrix is a matrix
Matrix most commonly refers to:
* ''The Matrix'' (franchise), an American media franchise
** ''The Matrix'', a 1999 science-fiction action film
** "The Matrix", a fictional setting, a virtual reality environment, within ''The Matrix'' (franchis ...
where each column and row represents one of the twenty standard amino acids. In bioinformatics
Bioinformatics () is an interdisciplinary field that develops methods and software tools for understanding biological data, in particular when the data sets are large and complex. As an interdisciplinary field of science, bioinformatics combi ...
, PAM matrices are sometimes used as substitution matrices to score sequence alignment
In bioinformatics, a sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity that may be a consequence of functional, structural, or evolutionary relationships between the sequences. Alig ...
s for proteins. Each entry in a PAM matrix indicates the likelihood of the amino acid of that row being replaced with the amino acid of that column through a series of one or more point accepted mutations during a specified evolutionary interval, rather than these two amino acids being aligned due to chance. Different PAM matrices correspond to different lengths of time in the evolution of the protein sequence.
Biological background
The genetic instructions of every replicatingcell
Cell most often refers to:
* Cell (biology), the functional basic unit of life
Cell may also refer to:
Locations
* Monastic cell, a small room, hut, or cave in which a religious recluse lives, alternatively the small precursor of a monastery ...
in a living organism are contained within its DNA. Throughout the cell's lifetime, this information is transcribed and replicated by cellular mechanisms to produce proteins or to provide instructions for daughter cells during cell division
Cell division is the process by which a parent cell (biology), cell divides into two daughter cells. Cell division usually occurs as part of a larger cell cycle in which the cell grows and replicates its chromosome(s) before dividing. In eukar ...
, and the possibility exists that the DNA may be altered during these processes. This is known as a mutation
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, mi ...
. At the molecular level, there are regulatory systems that correct most — but not all — of these changes to the DNA before it is replicated.
One of the possible mutations that occurs is the replacement of a single nucleotide
Nucleotides are organic molecules consisting of a nucleoside and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of which are essential biomolecules wi ...
, known as a point mutation. If a point mutation occurs within an expressed region of a gene
In biology, the word gene (from , ; "...Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a ba ...
, an exon
An exon is any part of a gene that will form a part of the final mature RNA produced by that gene after introns have been removed by RNA splicing. The term ''exon'' refers to both the DNA sequence within a gene and to the corresponding sequen ...
, then this will change the codon
The genetic code is the set of rules used by living cells to translate information encoded within genetic material ( DNA or RNA sequences of nucleotide triplets, or codons) into proteins. Translation is accomplished by the ribosome, which links ...
specifying a particular amino acid in the protein produced by that gene. Despite the redundancy in the genetic code
The genetic code is the set of rules used by living cells to translate information encoded within genetic material ( DNA or RNA sequences of nucleotide triplets, or codons) into proteins. Translation is accomplished by the ribosome, which links ...
, there is a possibility that this mutation will then change the amino acid that is produced during translation
Translation is the communication of the Meaning (linguistic), meaning of a #Source and target languages, source-language text by means of an Dynamic and formal equivalence, equivalent #Source and target languages, target-language text. The ...
, and as a consequence the structure of the protein will be changed.
The functionality of a protein is highly dependent on its structure. Changing a single amino acid in a protein may reduce its ability to carry out this function, or the mutation may even change the function that the protein carries out. Changes like these may severely impact a crucial function in a cell, potentially causing the cell — and in extreme cases, the organism — to die. Conversely, the change may allow the cell to continue functioning albeit differently, and the mutation can be passed on to the organism's offspring. If this change does not result in any significant physical disadvantage to the offspring, the possibility exists that this mutation will persist within the population. The possibility also exists that the change in function becomes advantageous. In either case, while being subjected to the processes of natural selection, the point mutation has been accepted into the genetic pool.
The 20 amino acids translated by the genetic code vary greatly by the physical and chemical properties of their side chains. However, these amino acids can be categorised into groups with similar physicochemical properties. Substituting an amino acid with another from the same category is more likely to have a smaller impact on the structure and function of a protein than replacement with an amino acid from a different category. Consequently, acceptance of point mutations depends heavily on the amino acid being replaced in the mutation, and the replacement amino acid. The PAM matrices are a mathematical tool that account for these varying rates of acceptance when evaluating the similarity of proteins during alignment.
Terminology
The term ''accepted point mutation'' was initially used to describe the mutation phenomenon. However, the acronym PAM was preferred over APM due to readability, and so the term ''point accepted mutation'' is used more regularly. Because the value in the PAMn matrix represents the number of mutations per 100 amino acids, which can be likened to a percentage of mutations, the term ''percentage accepted mutation'' is sometimes used. It is important to distinguish between point accepted mutations (PAMs), point accepted mutation matrices (PAM matrices) and the PAMn matrix. The term 'point accepted mutation' refers to the mutation event itself. However, 'PAM matrix' refers to one of a family of matrices which contain scores representing the likelihood of two amino acids being aligned due to a series of mutation events, rather than due to random chance. The 'PAMn matrix' is the PAM matrix corresponding to a time frame long enough for mutation events to occur per 100 amino acids.Construction of PAM matrices
PAM matrices were introduced byMargaret Dayhoff
Margaret Belle (Oakley) Dayhoff (March 11, 1925 – February 5, 1983) was an American physical chemist and a pioneer in the field of bioinformatics. Dayhoff was a professor at Georgetown University Medical Center and a noted research biochem ...
in 1978. The calculation of these matrices were based on 1572 observed mutations in the phylogenetic trees
A phylogenetic tree (also phylogeny or evolutionary tree Felsenstein J. (2004). ''Inferring Phylogenies'' Sinauer Associates: Sunderland, MA.) is a branching diagram or a tree showing the evolutionary relationships among various biological spec ...
of 71 families of closely related proteins. The proteins to be studied were selected on the basis of having high similarity with their predecessors. The protein alignments included were required to display at least 85% identity. As a result, it is reasonable to assume that any aligned mismatches were the result of a single mutation event, rather than several at the same location.
Each PAM matrix has twenty rows and twenty columns — one representing each of the twenty amino acids translated by the genetic code. The value in each cell of a PAM matrix is related to the probability of a row amino acid before the mutation being aligned with a column amino acid afterwards. From this definition, PAM matrices are an example of a substitution matrix
In bioinformatics and evolutionary biology, a substitution matrix describes the frequency at which a character in a nucleotide sequence or a protein sequence changes to other character states over evolutionary time. The information is often in ...
.
Collection of data from phylogenetic trees
For each branch in the phylogenetic trees of the protein families, the number of mismatches that were observed were recorded and a record kept of the two amino acids involved. These counts were used as entries below the main diagonal of the matrix . Since the vast majority of protein samples come from organisms that are alive today (extant species), the 'direction' of a mutation cannot be determined. That is, the amino acid present before the mutation cannot be distinguished from the amino acid that replaced it after the mutation. Because of this, the matrix is assumed to besymmetric
Symmetry (from grc, συμμετρία "agreement in dimensions, due proportion, arrangement") in everyday language refers to a sense of harmonious and beautiful proportion and balance. In mathematics, "symmetry" has a more precise definiti ...
, and the entries of above the main diagonal are computed on this basis. The entries along the diagonal of do not correspond to mutations and can be left unfilled.
In addition to these counts, data on the mutability and the frequency of the amino acids was obtained. The mutability of an amino acid is the ratio of the number of mutations it is involved in and the number of times it occurs in an alignment. Mutability measures how likely an amino acid is to mutate acceptably. Asparagine
Asparagine (symbol Asn or N) is an α-amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated −NH form under biological conditions), an α-carboxylic acid group (which is in the depro ...
, an amino acid with a small polar
Polar may refer to:
Geography
Polar may refer to:
* Geographical pole, either of two fixed points on the surface of a rotating body or planet, at 90 degrees from the equator, based on the axis around which a body rotates
* Polar climate, the c ...
side chain, was found to be the most mutable of the amino acids. Cysteine
Cysteine (symbol Cys or C; ) is a semiessential proteinogenic amino acid with the formula . The thiol side chain in cysteine often participates in enzymatic reactions as a nucleophile.
When present as a deprotonated catalytic residue, sometime ...
and tryptophan
Tryptophan (symbol Trp or W)
is an α-amino acid that is used in the biosynthesis of proteins. Tryptophan contains an α-amino group, an α- carboxylic acid group, and a side chain indole, making it a polar molecule with a non-polar aromatic ...
were found to be the least mutable amino acids. The side chains for cysteine and tryptophan have less common structures: cysteine's side chain contains sulfur which participates in disulfide bonds
In biochemistry, a disulfide (or disulphide in British English) refers to a functional group with the structure . The linkage is also called an SS-bond or sometimes a disulfide bridge and is usually derived by the coupling of two thiol groups. In ...
with other cysteine molecules, and tryptophan's side chain is large and aromatic
In chemistry, aromaticity is a chemical property of cyclic ( ring-shaped), ''typically'' planar (flat) molecular structures with pi bonds in resonance (those containing delocalized electrons) that gives increased stability compared to satur ...
. Since there are several small polar amino acids, these extremes suggest that amino acids are more likely to acceptably mutate if their physical and chemical properties are more common among alternative amino acids.
Construction of the mutation matrix
For the th amino acid, the values and are its mutability and frequency. The frequencies of the amino acids are normalised so that they sum to 1. If total number of occurrences of the th amino acid is , and is the total number of all amino acids, then : Based on the definition of mutability as the ratio of mutations to occurrences of an amino acid : or : The mutation matrix is constructed so that the entry represents the probability of the th amino acid mutating into the th amino acid. The non-diagonal entries are computed by the equation : where is a constant of proportionality. However, this equation does not compute the diagonal entries. Each column in the matrix lists each of the twenty possible outcomes for an amino acid — it can mutate into one of the 19 other amino acids, or remain unchanged. Since the non-diagonal entries listing the probabilities of each of the 19 mutations are known, and the sum of the probabilities of these twenty outcomes must be 1, this last probability can be calculated by : which simplifies to : : A result of particular significance is that for the non-diagonal entries : Which means that for all entries in the mutation matrix :Choice of the constant of proportionality
The probabilities contained in vary as some unknown function of the amount of time that a protein sequence is allowed to mutate for. Instead of attempting to determine this relationship, the values of are calculated for a short time frame, and the matrices for longer periods of time are calculated by assuming mutations follow aMarkov chain
A Markov chain or Markov process is a stochastic model describing a sequence of possible events in which the probability of each event depends only on the state attained in the previous event. Informally, this may be thought of as, "What happe ...
model. The base unit of time for the PAM matrices is the time required for 1 mutation to occur per 100 amino acids, sometimes called 'a PAM unit' or 'a PAM' of time. This is precisely the duration of mutation assumed by the PAM1 matrix.
The constant is used to control the proportion of amino acids that are unchanged. By using only alignments of proteins that had at least 85% similarity, it could be reasonably assumed that the mutations observed were direct, without any intermediate states. This means that scaling down these counts by a common factor would provide an accurate estimate of the mutation counts had the similarity been closer to 100%. It also means that the number of mutations per 100 amino acids, the in PAMn is equal to the number of mutated amino acids per 100 amino acids.
To find the mutation matrix for the PAM1 matrix, the requirement that 99% of the amino acids in a sequence are conserved is imposed. The quantity is equal to the number of conserved amino acid units, and so the total number of conserved amino acids is
:
The value of needed to be pick to produce 99% identity after mutation is then given by the equation
:
This value can then be used in the mutation matrix for the PAM1 matrix.
Construction of the PAMn matrices
The Markov chain model of protein mutation relates the mutation matrix for PAMn, , to the mutation matrix for the PAM1 matrix, by the simple relationship : The PAMn matrix is constructed from the ratio of the probability of point accepted mutations replacing the th amino acid with the th amino acid, to the probability of these amino acids being aligned by chance. The entries of the PAMn matrix are given by the equation : Note that in Gusfield's book, the entries and are related to the probability of the th amino acid mutating into the th amino acid. This is the origin of the different equation for the entries of the PAM matrices. When using the PAMn matrix to score an alignment of two proteins, the following assumption is made: ::''If these two proteins are related, the evolutionary interval separating them is the time taken for point accepted mutations to occur per 100 amino acids.'' When the alignment of the th and th amino acids is considered, the score indicates the relative likelihoods of the alignment due to the proteins being related or due to random chance. * If the proteins are related, a series of point accepted mutations must have occurred to mutate the original amino acid into its replacement. Suppose the th amino acid is the original. Based on the abundance of amino acids in proteins, the probability of the th amino acid being the original is . Given any particular unit of this amino acid, theprobability
Probability is the branch of mathematics concerning numerical descriptions of how likely an Event (probability theory), event is to occur, or how likely it is that a proposition is true. The probability of an event is a number between 0 and ...
of being replaced by the th amino acid in the assumed time interval is . Thus, the probability of the alignment is , the numerator within the logarithm.
* If the proteins are not related, the events that the two aligned amino acids are the th and th amino acids must be independent
Independent or Independents may refer to:
Arts, entertainment, and media Artist groups
* Independents (artist group), a group of modernist painters based in the New Hope, Pennsylvania, area of the United States during the early 1930s
* Independ ...
. The probabilities of these events are and , which means the probability of the alignment is , the denominator of the logarithm.
* Thus, the logarithm in the equation results in a positive entry if the alignment is more likely due to point accepted mutations, and a negative entry if the alignment is more likely due to chance.
Properties of the PAM matrices
Symmetry of the PAM matrices
While the mutation probability matrix is not symmetric, each of the PAM matrices are. This somewhat surprising property is a result of the relationship that was noted for the mutation probability matrix: : In fact, this relationship holds for all positive integer powers of the matrix : : : As a result, the entries of the PAMn matrix are symmetric, since :Relating the number of mutated amino acids and the number of mutations
The value represents the number of mutations that occur per 100 amino acids, however this value is rarely accessible and often estimated. However, when comparing two proteins it is easy to calculate instead, which is the number of mutated amino acids per 100 amino acids. Despite the random nature of mutation, these values can be approximately related by : : The validity of these estimates can be verified by counting the number of amino acids that remain unchanged under the action of the matrix . The total number of unchanged amino acids for the time interval of the PAMn matrix is : and so the proportion of unchanged amino acids is :An example - PAM250
A PAM250 is a commonly used scoring matrix for sequence comparison. Only the lower half of the matrix needs to be computed, since by their construction, PAM matrices are required to be symmetric. Each of the 20 amino acid are shown down the top and side of the matrix, with 3 additional ambiguous amino acids. The amino acids are most commonly shown listed alphabetically, or listed in groups. Thesegroups
A group is a number of persons or things that are located, gathered, or classed together.
Groups of people
* Cultural group, a group whose members share the same cultural identity
* Ethnic group, a group whose members share the same ethnic ide ...
are the characteristics shared among the amino acids.

Uses in bioinformatics
Determining the time of divergence in phylogenetic trees
The molecular clock hypothesis predicts that the rate of amino acid substitution in a particular protein will be approximately constant over time, though this rate may vary between protein families. This suggests that the number of mutations per amino acid in a protein increases approximately linearly with time. Determining the time at which two proteins diverged is an important task inphylogenetics
In biology, phylogenetics (; from Greek language, Greek wikt:φυλή, φυλή/wikt:φῦλον, φῦλον [] "tribe, clan, race", and wikt:γενετικός, γενετικός [] "origin, source, birth") is the study of the evolutionary his ...
. Fossil#Estimating dates, Fossil records are often used to establish the position of events on the timeline of the Earth's evolutionary history, but the application of this source is limited. However, if the rate at which the molecular clock of protein family ticks — that is, the rate at which the number of mutations per amino acid increases — is known, then knowing this number of mutations would allow the date of divergence to be found.
Suppose the date of divergence for two related proteins, taken from organisms living today, is sought. The two proteins have both been accumulating accepted mutations since the date of divergence, and so the total number of mutations per amino acid separating them is approximately twice that which separates them from their common ancestor
Common descent is a concept in evolutionary biology applicable when one species is the ancestor of two or more species later in time. All living beings are in fact descendants of a unique ancestor commonly referred to as the last universal comm ...
. If a range of PAM matrices are used to align two proteins that are known to be related, then the value of in the PAMn matrix which results in the best score is most likely to correspond to the mutations per amino acid separating the two proteins. Halving this value and dividing by the rate at which accepted mutations accumulate in the protein family provides an estimate of the time of divergence of these two proteins from their common ancestor. That is, the time of divergence in myr is
:
Where is the number of mutations per amino acid, and is the rate of accepted mutation accumulation in mutations per amino acid site per million years.
Use in BLAST
PAM matrices are also used as a scoring matrix when comparing DNA sequences or protein sequences to judge the quality of the alignment. This form of scoring system is utilized by a wide range of alignment software includingBLAST
Blast or The Blast may refer to:
*Explosion, a rapid increase in volume and release of energy in an extreme manner
*Detonation, an exothermic front accelerating through a medium that eventually drives a shock front
Film
* ''Blast'' (1997 film), ...
.
Comparing PAM and BLOSUM
Although the PAM log-odds matrices were the first scoring matrices used with BLAST, the PAM matrices have largely been replaced by theBLOSUM
In bioinformatics, the BLOSUM (BLOcks SUbstitution Matrix) matrix is a substitution matrix used for sequence alignment of proteins. BLOSUM matrices are used to score alignments between evolutionarily divergent protein sequences. They are based o ...
matrices. Although both matrices produce similar scoring outcomes they were generated using differing methodologies. The BLOSUM matrices were generated directly from the amino acid differences in aligned blocks that have diverged to varying degrees the PAM matrices reflect the extrapolation
In mathematics, extrapolation is a type of estimation, beyond the original observation range, of the value of a variable on the basis of its relationship with another variable. It is similar to interpolation, which produces estimates between know ...
of evolutionary information based on closely related sequences to longer timescales. Since scoring information for the PAM and BLOSUM matrices were generated in very different ways the numbers associated with the matrices have fundamentally different meanings; the numbers for PAM matrices increase for comparisons among more divergent proteins whereas the numbers for the BLOSUM matrices decrease. However, all amino acid substitution matrices can be compared in an information theoretic framework using their relative entropy.
See also
*Point mutation
A point mutation is a genetic mutation where a single nucleotide base is changed, inserted or deleted from a DNA or RNA sequence of an organism's genome. Point mutations have a variety of effects on the downstream protein product—consequences ...
* Sequence alignment
In bioinformatics, a sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity that may be a consequence of functional, structural, or evolutionary relationships between the sequences. Alig ...
* Margaret Dayhoff
Margaret Belle (Oakley) Dayhoff (March 11, 1925 – February 5, 1983) was an American physical chemist and a pioneer in the field of bioinformatics. Dayhoff was a professor at Georgetown University Medical Center and a noted research biochem ...
* Molecular clock
The molecular clock is a figurative term for a technique that uses the mutation rate of biomolecules to deduce the time in prehistory when two or more life forms diverged. The biomolecular data used for such calculations are usually nucleoti ...
* BLOSUM
In bioinformatics, the BLOSUM (BLOcks SUbstitution Matrix) matrix is a substitution matrix used for sequence alignment of proteins. BLOSUM matrices are used to score alignments between evolutionarily divergent protein sequences. They are based o ...
* BLAST
Blast or The Blast may refer to:
*Explosion, a rapid increase in volume and release of energy in an extreme manner
*Detonation, an exothermic front accelerating through a medium that eventually drives a shock front
Film
* ''Blast'' (1997 film), ...
References
{{reflist, 2External links
* http://www.inf.ethz.ch/personal/gonnet/DarwinManual/node148.html * http://www.bioinformatics.nl/tools/pam.html For quickly calculating a PAM matrix. * http://web.expasy.org/docs/relnotes/relstat.html The most recent statistics from the Swiss-Prot protein knowledgebase. Section 6.1 contains the most up-to-date amino acid frequencies Mutation Bioinformatics