Non-canonical base pairing on:
[Wikipedia]
[Google]
[Amazon]
Non-canonical base pairing occurs when nucleobases hydrogen bond, or
 An estimated 60% of bases in structured RNA participate in canonical Watson-Crick base pairs. Base pairing occurs when two bases form hydrogen bonds with each other. These hydrogen bonds can be either polar or non-polar interactions. The polar hydrogen bonds are formed by N-H...O/N and/or O-H...O/N interactions. Non-polar hydrogen bonds are formed between C-H...O/N.
An estimated 60% of bases in structured RNA participate in canonical Watson-Crick base pairs. Base pairing occurs when two bases form hydrogen bonds with each other. These hydrogen bonds can be either polar or non-polar interactions. The polar hydrogen bonds are formed by N-H...O/N and/or O-H...O/N interactions. Non-polar hydrogen bonds are formed between C-H...O/N.
 Besides the three edges of interaction, base pairs can also vary in their cis/trans forms. The cis and trans structures depend on the orientation of the ribose sugar as opposed to the hydrogen bond interaction. These various orientations are shown in Figure 3. Therefore, with the cis/trans forms and the 3 hydrogen bond edges, there are 12 basic types of base pairing geometries which can be found in RNA structures. Those 12 types are WC:WC (cis/trans), W:HC (cis/trans), WC:S (cis/trans), H:S (cis/trans), H:H (cis/trans), and S:S (cis/trans).
Besides the three edges of interaction, base pairs can also vary in their cis/trans forms. The cis and trans structures depend on the orientation of the ribose sugar as opposed to the hydrogen bond interaction. These various orientations are shown in Figure 3. Therefore, with the cis/trans forms and the 3 hydrogen bond edges, there are 12 basic types of base pairing geometries which can be found in RNA structures. Those 12 types are WC:WC (cis/trans), W:HC (cis/trans), WC:S (cis/trans), H:S (cis/trans), H:H (cis/trans), and S:S (cis/trans).
 The bonds between various bases are well defined because of their rigid and planar shape. The spatial interactions between the two bases can be classified in 6 rigid-body parameters or intra-base pair parameters (3 translational, 3 rotational) as shown in Figure 4. These parameters describe the base pairs' three dimensional conformation.
The three translational arrangements are known as shear, stretch, and stagger. These three parameters are directly related to the proximity and direction of the hydrogen bonds. The rotational arrangements are buckle, propeller, and opening. Rotational arrangements relate to the non-planar confirmation (as compared to the ideal coplanar geometry). Intra-base pair parameters are used to determine the structure and stabilities of non-canonical base pairs and were originally created for the base pairings in DNA, but were found to also fit the non-canonical base models.
The bonds between various bases are well defined because of their rigid and planar shape. The spatial interactions between the two bases can be classified in 6 rigid-body parameters or intra-base pair parameters (3 translational, 3 rotational) as shown in Figure 4. These parameters describe the base pairs' three dimensional conformation.
The three translational arrangements are known as shear, stretch, and stagger. These three parameters are directly related to the proximity and direction of the hydrogen bonds. The rotational arrangements are buckle, propeller, and opening. Rotational arrangements relate to the non-planar confirmation (as compared to the ideal coplanar geometry). Intra-base pair parameters are used to determine the structure and stabilities of non-canonical base pairs and were originally created for the base pairings in DNA, but were found to also fit the non-canonical base models.


 Basic secondary structural elements of RNA include bulges, double helices, hairpin loops, and internal loops. An example of a hairpin loop of RNA is given in Figure 7. As shown in the figure, hairpin loops and internal loops require a sudden change in backbone direction. Non-canonical base pairing allows for the increased flexibility at junctions or turns required in the secondary structure.
Basic secondary structural elements of RNA include bulges, double helices, hairpin loops, and internal loops. An example of a hairpin loop of RNA is given in Figure 7. As shown in the figure, hairpin loops and internal loops require a sudden change in backbone direction. Non-canonical base pairing allows for the increased flexibility at junctions or turns required in the secondary structure.
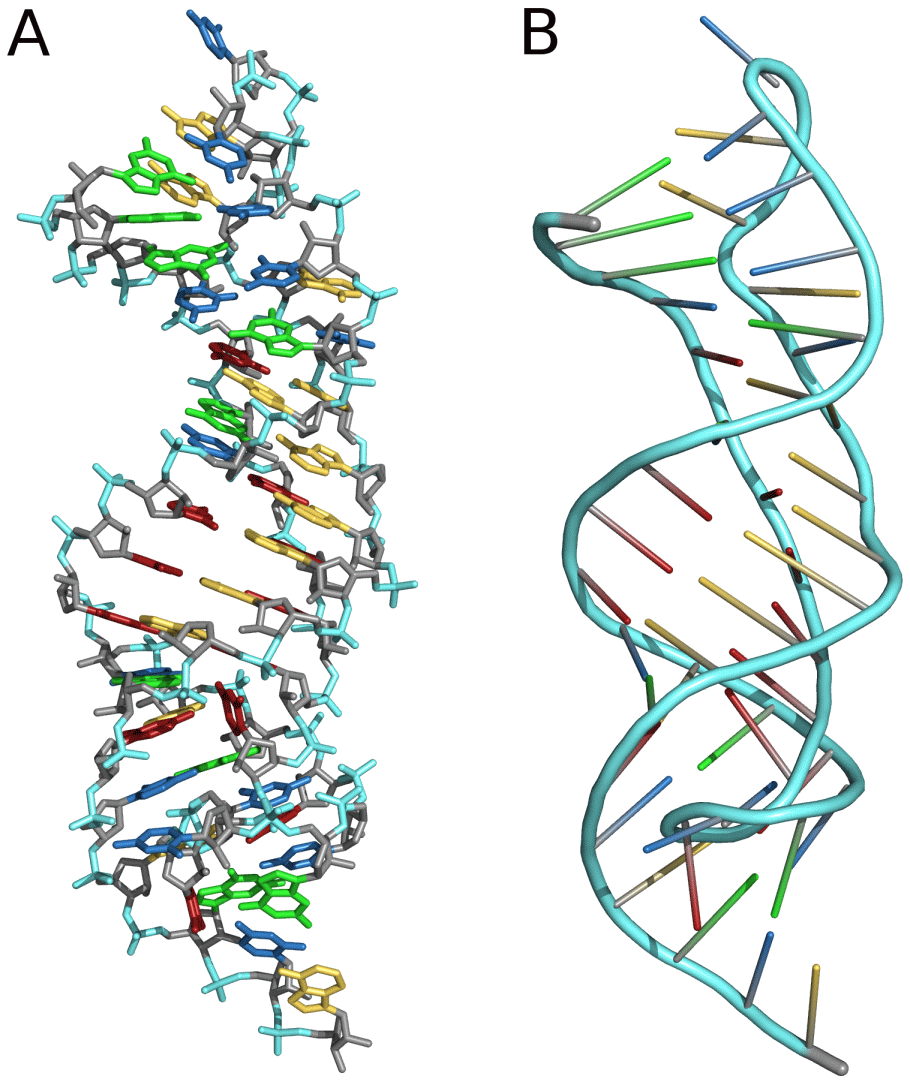 Three-dimensional structures are formed through the long-range intra-molecular interactions between the secondary structures. This leads to the formation of pseudoknots, ribose zippers, kissing hairpin loops, or co-axial pseudocontinuous helices. The three-dimensional structures of RNA are primarily determined through molecular simulations or computationally guided measurements. An example of a Pseudoknot is given in Figure 8.
Three-dimensional structures are formed through the long-range intra-molecular interactions between the secondary structures. This leads to the formation of pseudoknots, ribose zippers, kissing hairpin loops, or co-axial pseudocontinuous helices. The three-dimensional structures of RNA are primarily determined through molecular simulations or computationally guided measurements. An example of a Pseudoknot is given in Figure 8.
base pair
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA ...
, to one another in schemes other than the standard Watson-Crick base pairs (which are adenine
Adenine () (symbol A or Ade) is a nucleobase (a purine derivative). It is one of the four nucleobases in the nucleic acid of DNA that are represented by the letters G–C–A–T. The three others are guanine, cytosine and thymine. Its derivativ ...
(A) -- thymine
Thymine () (symbol T or Thy) is one of the four nucleobases in the nucleic acid of DNA that are represented by the letters G–C–A–T. The others are adenine, guanine, and cytosine. Thymine is also known as 5-methyluracil, a pyrimidine nuc ...
(T) in DNA, adenine (A) -- uracil (U) in RNA, and guanine
Guanine () (symbol G or Gua) is one of the four main nucleobases found in the nucleic acids DNA and RNA, the others being adenine, cytosine, and thymine (uracil in RNA). In DNA, guanine is paired with cytosine. The guanine nucleoside is called ...
(G) -- cytosine
Cytosine () (symbol C or Cyt) is one of the four nucleobases found in DNA and RNA, along with adenine, guanine, and thymine (uracil in RNA). It is a pyrimidine derivative, with a heterocyclic aromatic ring and two substituents attached (an amin ...
(C) in both DNA and RNA). There are three main types of non-canonical base pairs: those stabilized by polar hydrogen bonds, those having interactions among C−H and O/N groups, and those that have hydrogen bonds between the bases themselves. The first discovered non-canonical base pairs are Hoogsteen base pairs, which were first described by American biochemist Karst Hoogsteen
Karst Hoogsteen (October 1, 1923 – August 10, 2015) was a Dutch-born American biochemist famous for noting a new base pairing form in DNA, now called Hoogsteen base pairs.. These base pairing intercede in the Watson-Crick base pairing
A base ...
.
Non-canonical base pairings commonly occur in the secondary structure of RNA (e.g. pairing of G with U), and in tRNA
Transfer RNA (abbreviated tRNA and formerly referred to as sRNA, for soluble RNA) is an adaptor molecule composed of RNA, typically 76 to 90 nucleotides in length (in eukaryotes), that serves as the physical link between the mRNA and the amino a ...
recognition. They are typically less stable than standard base pairings. The presence of non-canonical base pairs in double stranded DNA results in a disrupted double helix.
History
James Watson and Francis Crick published the double helical structure of DNA and proposed the canonical Watson-Crick base pairs in 1953. Ten years later, in 1963,Karst Hoogsteen
Karst Hoogsteen (October 1, 1923 – August 10, 2015) was a Dutch-born American biochemist famous for noting a new base pairing form in DNA, now called Hoogsteen base pairs.. These base pairing intercede in the Watson-Crick base pairing
A base ...
reported that he had used single crystal X-ray diffraction to investigate alternative base pair structures, and he found an alternative structure for the nucelobase pair adenine-thymine in which the purine (A) takes on an alternative conformation with respect to the pyrimidine
Pyrimidine (; ) is an aromatic, heterocyclic, organic compound similar to pyridine (). One of the three diazines (six-membered heterocyclics with two nitrogen atoms in the ring), it has nitrogen atoms at positions 1 and 3 in the ring. The othe ...
(T). Five years after Hoogsteen proposed the A-T Hoogsteen base pair
A Hoogsteen base pair is a variation of base-pairing in nucleic acids such as the A•T pair. In this manner, two nucleobases, one on each strand, can be held together by hydrogen bonds in the major groove. A Hoogsteen base pair applies the N7 pos ...
, optical rotary dispersion spectra which provided evidence for a G-C Hoogsteen base pair were reported. The G-C Hoogsteen base pair was first observed via X-ray crystallography years later, in 1986, by co-crystallizing DNA with triostin A (an antibiotic
An antibiotic is a type of antimicrobial substance active against bacteria. It is the most important type of antibacterial agent for fighting bacterial infections, and antibiotic medications are widely used in the treatment and prevention ...
). Ultimately, after years of studying both Watson-Crick and Hoogsteen base pairs, it has been determined that both occur naturally in DNA, and that they exist in equilibrium with one another; the conditions in which the DNA exists ultimately determine which form will be favored. Since the structures of the canonical Watson-Crick and non-canonical Hoogsteen base pairs were determined, many other types of non-canonical base pairs have been presented and described.
Structure
Base pairing
 An estimated 60% of bases in structured RNA participate in canonical Watson-Crick base pairs. Base pairing occurs when two bases form hydrogen bonds with each other. These hydrogen bonds can be either polar or non-polar interactions. The polar hydrogen bonds are formed by N-H...O/N and/or O-H...O/N interactions. Non-polar hydrogen bonds are formed between C-H...O/N.
An estimated 60% of bases in structured RNA participate in canonical Watson-Crick base pairs. Base pairing occurs when two bases form hydrogen bonds with each other. These hydrogen bonds can be either polar or non-polar interactions. The polar hydrogen bonds are formed by N-H...O/N and/or O-H...O/N interactions. Non-polar hydrogen bonds are formed between C-H...O/N.
Edge interactions
Each base has three potential edges where it can interact with another base. The Purine bases have 3 edges which are able to hydrogen bond. Those are known as the Watson-Crick edge(WC), the Hoogsteen edge(H), and the Sugar edge(S). Pyrimidine bases also have three hydrogen-bonding edges. Like the purine, there is the Watson-Crick edge(WC) and the Sugar edge(S) but the third edge is referred to as the "C-H" edge(H) on the pyrimidine bases. This C-H edge is sometimes also referred to as the Hoogsteen edge for simplicity. The various edges for the purine and pyrimidine bases are shown in Figure 2. Besides the three edges of interaction, base pairs can also vary in their cis/trans forms. The cis and trans structures depend on the orientation of the ribose sugar as opposed to the hydrogen bond interaction. These various orientations are shown in Figure 3. Therefore, with the cis/trans forms and the 3 hydrogen bond edges, there are 12 basic types of base pairing geometries which can be found in RNA structures. Those 12 types are WC:WC (cis/trans), W:HC (cis/trans), WC:S (cis/trans), H:S (cis/trans), H:H (cis/trans), and S:S (cis/trans).
Besides the three edges of interaction, base pairs can also vary in their cis/trans forms. The cis and trans structures depend on the orientation of the ribose sugar as opposed to the hydrogen bond interaction. These various orientations are shown in Figure 3. Therefore, with the cis/trans forms and the 3 hydrogen bond edges, there are 12 basic types of base pairing geometries which can be found in RNA structures. Those 12 types are WC:WC (cis/trans), W:HC (cis/trans), WC:S (cis/trans), H:S (cis/trans), H:H (cis/trans), and S:S (cis/trans).
Classification
These 12 types can be further divided into more subgroups which are dependent on the directionality of the glycosidic bonds and steric extensions. With all of the various base pair combinations there are 169 theoretically possible base pair combinations. The actual number ofbase-pair
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA ...
combinations is lower because some combinations result in non-favorable interactions. This number of possible non-canonical base pairs is still being determined as it depends strongly on base pairing criteria . Understanding base pair configuration is similarly difficult since the pairing is depends on the bases surroundings. These surroundings can consist of adjacent base pairs, adjacent loops, or third interactions (such as a base triple).
 The bonds between various bases are well defined because of their rigid and planar shape. The spatial interactions between the two bases can be classified in 6 rigid-body parameters or intra-base pair parameters (3 translational, 3 rotational) as shown in Figure 4. These parameters describe the base pairs' three dimensional conformation.
The three translational arrangements are known as shear, stretch, and stagger. These three parameters are directly related to the proximity and direction of the hydrogen bonds. The rotational arrangements are buckle, propeller, and opening. Rotational arrangements relate to the non-planar confirmation (as compared to the ideal coplanar geometry). Intra-base pair parameters are used to determine the structure and stabilities of non-canonical base pairs and were originally created for the base pairings in DNA, but were found to also fit the non-canonical base models.
The bonds between various bases are well defined because of their rigid and planar shape. The spatial interactions between the two bases can be classified in 6 rigid-body parameters or intra-base pair parameters (3 translational, 3 rotational) as shown in Figure 4. These parameters describe the base pairs' three dimensional conformation.
The three translational arrangements are known as shear, stretch, and stagger. These three parameters are directly related to the proximity and direction of the hydrogen bonds. The rotational arrangements are buckle, propeller, and opening. Rotational arrangements relate to the non-planar confirmation (as compared to the ideal coplanar geometry). Intra-base pair parameters are used to determine the structure and stabilities of non-canonical base pairs and were originally created for the base pairings in DNA, but were found to also fit the non-canonical base models.
Types
The most common non-canonical base pairs are trans A:G Hoogsteen/sugar edge, A:U Hoogsteen/WC, and G:U Wobble pairs.Hoogsteen base pairs
Hoogsteen base pair
A Hoogsteen base pair is a variation of base-pairing in nucleic acids such as the A•T pair. In this manner, two nucleobases, one on each strand, can be held together by hydrogen bonds in the major groove. A Hoogsteen base pair applies the N7 pos ...
s occur between adenine (A) and thymine (T); and guanine (G) and cytosine(C); similarly to Watson-Crick base pairs. However, the purine (A and G) takes on an alternative conformation with respect to the pyrimidine
Pyrimidine (; ) is an aromatic, heterocyclic, organic compound similar to pyridine (). One of the three diazines (six-membered heterocyclics with two nitrogen atoms in the ring), it has nitrogen atoms at positions 1 and 3 in the ring. The othe ...
. In the A-U Hoogsteen base pair, the adenine is rotated 180° about the glycosidic bond, resulting in an alternative hydrogen bonding scheme which has one hydrogen bond in common with the Watson-Crick base pair (adenine N6 and thymine N4), while the other hydrogen bond, instead of occurring between adenine N1 and thymine N3 as in the Watson-Crick base pair, occurs between adenine N7 and thymine N3. The A-U base pair is shown in Figure 5. In the G-C Watson-Crick base pair, like the A-T Hoogsteen base pair, the purine (guanine) is rotated 180° about the glycosidic bond while the pyrimidine (cytosine) remains in place. One hydrogen bond from the Watson-Crick base pair is maintained (guanine O6 and cytosine N4) and the other occurs between guanine N7 and a protonated cytosine N3 (note that the Hoogsteen G-C base pair has two hydrogen bonds, while the Watson-Crick G-C base pair has three).
Wobble base pairs
Wobble base pairing occur between two nucleotides that are not Watson-Crick base pairs and was proposed byWatson
Watson may refer to:
Companies
* Actavis, a pharmaceutical company formerly known as Watson Pharmaceuticals
* A.S. Watson Group, retail division of Hutchison Whampoa
* Thomas J. Watson Research Center, IBM research center
* Watson Systems, make ...
in 1966. The 4 main examples are guanine-uracil (G-U), hypoxanthine-uracil (I-U), hypoxanthine-adenine (I-A), and hypoxanthine-cytosine (I-C). These wobble base pairs are very important in tRNA. Most organisms have less than 45 tRNA molecules even though 61 tRNA molecules would technically be necessary to canonically pair to the codon. Wobble base pairing allows for the 5' anticodon to bond to a non-standard base pair. Examples of wobble base pairs are given in Figure 6.
3-D Structure
The secondary and three-dimensional structures of RNA are formed and stabilized through non-canonical base pairs. Base pairs make up many secondary structural blocks which aid the folding of RNA complexes and three dimensional structures. The overall folded RNA is stabilized by the tertiary and secondary structures canonically base pairing together. Due to the many possible non-canonical base pairs, there are an unlimited amount of structures, which allows for the diverse functions of RNA. The arrangement of the non-canonical bases also allow long-range RNA interactions, recognition of proteins and other molecules, and structural stabilizing elements. Many of the common non-canonical base pairs can be added to a stacked RNA stem without disturbing its helical character.Secondary
 Basic secondary structural elements of RNA include bulges, double helices, hairpin loops, and internal loops. An example of a hairpin loop of RNA is given in Figure 7. As shown in the figure, hairpin loops and internal loops require a sudden change in backbone direction. Non-canonical base pairing allows for the increased flexibility at junctions or turns required in the secondary structure.
Basic secondary structural elements of RNA include bulges, double helices, hairpin loops, and internal loops. An example of a hairpin loop of RNA is given in Figure 7. As shown in the figure, hairpin loops and internal loops require a sudden change in backbone direction. Non-canonical base pairing allows for the increased flexibility at junctions or turns required in the secondary structure.
Tertiary
 Three-dimensional structures are formed through the long-range intra-molecular interactions between the secondary structures. This leads to the formation of pseudoknots, ribose zippers, kissing hairpin loops, or co-axial pseudocontinuous helices. The three-dimensional structures of RNA are primarily determined through molecular simulations or computationally guided measurements. An example of a Pseudoknot is given in Figure 8.
Three-dimensional structures are formed through the long-range intra-molecular interactions between the secondary structures. This leads to the formation of pseudoknots, ribose zippers, kissing hairpin loops, or co-axial pseudocontinuous helices. The three-dimensional structures of RNA are primarily determined through molecular simulations or computationally guided measurements. An example of a Pseudoknot is given in Figure 8.
Experimental Methods
Watson-Crick canonical base pairing is not the only edge-to-edge conformation possible for the nucleotide since non-canonical pairing can take place as well. Sugar-phosphate backbone has an ionic character, which makes the bases sensitive to their environment, leading to conformational changes, such as non-canonical pairing. There are various methods of prediction for these conformations, such asNMR
Nuclear magnetic resonance (NMR) is a physical phenomenon in which nuclei in a strong constant magnetic field are perturbed by a weak oscillating magnetic field (in the near field) and respond by producing an electromagnetic signal with a ...
structure determination and X-ray crystallography.
Applications
RNA has a multitude of purposes throughout the cell including regulating many important steps in gene expression. Various conformations of the non-Watson-Crick base pairs allow for a multitude of biological functions such as mRNA splicing, siRNA, transport, protein recognition, protein binding, and translation. One common example of a biological application of non-canonical base pairs is thekink turn
Kink or KINK may refer to:
Common uses
* Kink (sexuality), a colloquial term for non-normative sexual behavior
* Kink, a curvature, bend, or twist
Geography
* Kink, Iran, a village in Iran
* The Kink, a man-made geographic feature in remote ea ...
. A kink-turn is found throughout many functional RNA species. It consists of a three-nucleotide bulge due to three Hoogsteen base pairs. This kink-turn acts as a marker where various proteins such as the human 15-5k protein or proteins in the L7Ae family can bind. A similar scenario is described in the binding of the HIV-1 Rev-response element (RRE) RNA. RRE RNA has an extra wide deep groove that is caused by cis Watson-Crick G:A pair followed by a trans Watson-Crick G:G. The HIV-1 Rev-response element is then able to bind due to the deepened groove.
See also
*Hoogsteen base pair
A Hoogsteen base pair is a variation of base-pairing in nucleic acids such as the A•T pair. In this manner, two nucleobases, one on each strand, can be held together by hydrogen bonds in the major groove. A Hoogsteen base pair applies the N7 pos ...
* Wobble base pair
References
{{reflist Molecular genetics Nucleic acids