In Situ Hybridization, Fluorescence on:
[Wikipedia]
[Google]
[Amazon]

 Fluorescence ''in situ'' hybridization (FISH) is a molecular cytogenetic technique that uses
Fluorescence ''in situ'' hybridization (FISH) is a molecular cytogenetic technique that uses
 In biology, a probe is a single strand of DNA or RNA that is complementary to a nucleotide sequence of interest.
RNA probes can be designed for any gene or any sequence within a gene for visualization of mRNA, lncRNA and miRNA in tissues and cells. FISH is used by examining the cellular reproduction cycle, specifically interphase of the nuclei for any chromosomal abnormalities. FISH allows the analysis of a large series of archival cases much easier to identify the pinpointed chromosome by creating a probe with an artificial chromosomal foundation that will attract similar chromosomes. The hybridization signals for each probe when a nucleic abnormality is detected. Each probe for the detection of mRNA and lncRNA is composed of ~20-50 oligonucleotide pairs, each pair covering a space of 40–50 bp. The specifics depend on the specific FISH technique used. For miRNA detection, the probes use proprietary chemistry for specific detection of miRNA and cover the entire miRNA sequence.
In biology, a probe is a single strand of DNA or RNA that is complementary to a nucleotide sequence of interest.
RNA probes can be designed for any gene or any sequence within a gene for visualization of mRNA, lncRNA and miRNA in tissues and cells. FISH is used by examining the cellular reproduction cycle, specifically interphase of the nuclei for any chromosomal abnormalities. FISH allows the analysis of a large series of archival cases much easier to identify the pinpointed chromosome by creating a probe with an artificial chromosomal foundation that will attract similar chromosomes. The hybridization signals for each probe when a nucleic abnormality is detected. Each probe for the detection of mRNA and lncRNA is composed of ~20-50 oligonucleotide pairs, each pair covering a space of 40–50 bp. The specifics depend on the specific FISH technique used. For miRNA detection, the probes use proprietary chemistry for specific detection of miRNA and cover the entire miRNA sequence.
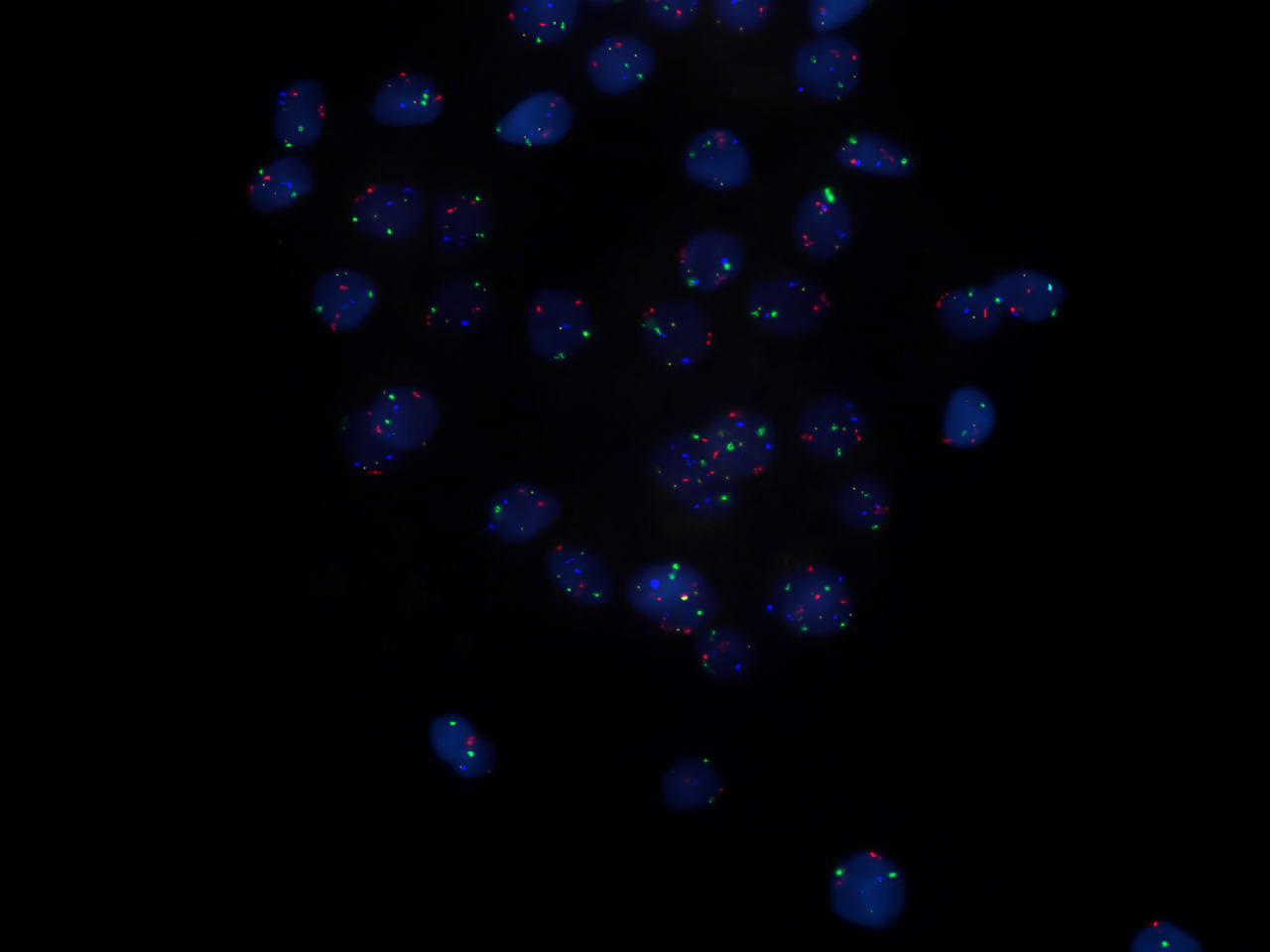 Probes are often derived from fragments of DNA that were isolated, purified, and amplified for use in the
Probes are often derived from fragments of DNA that were isolated, purified, and amplified for use in the
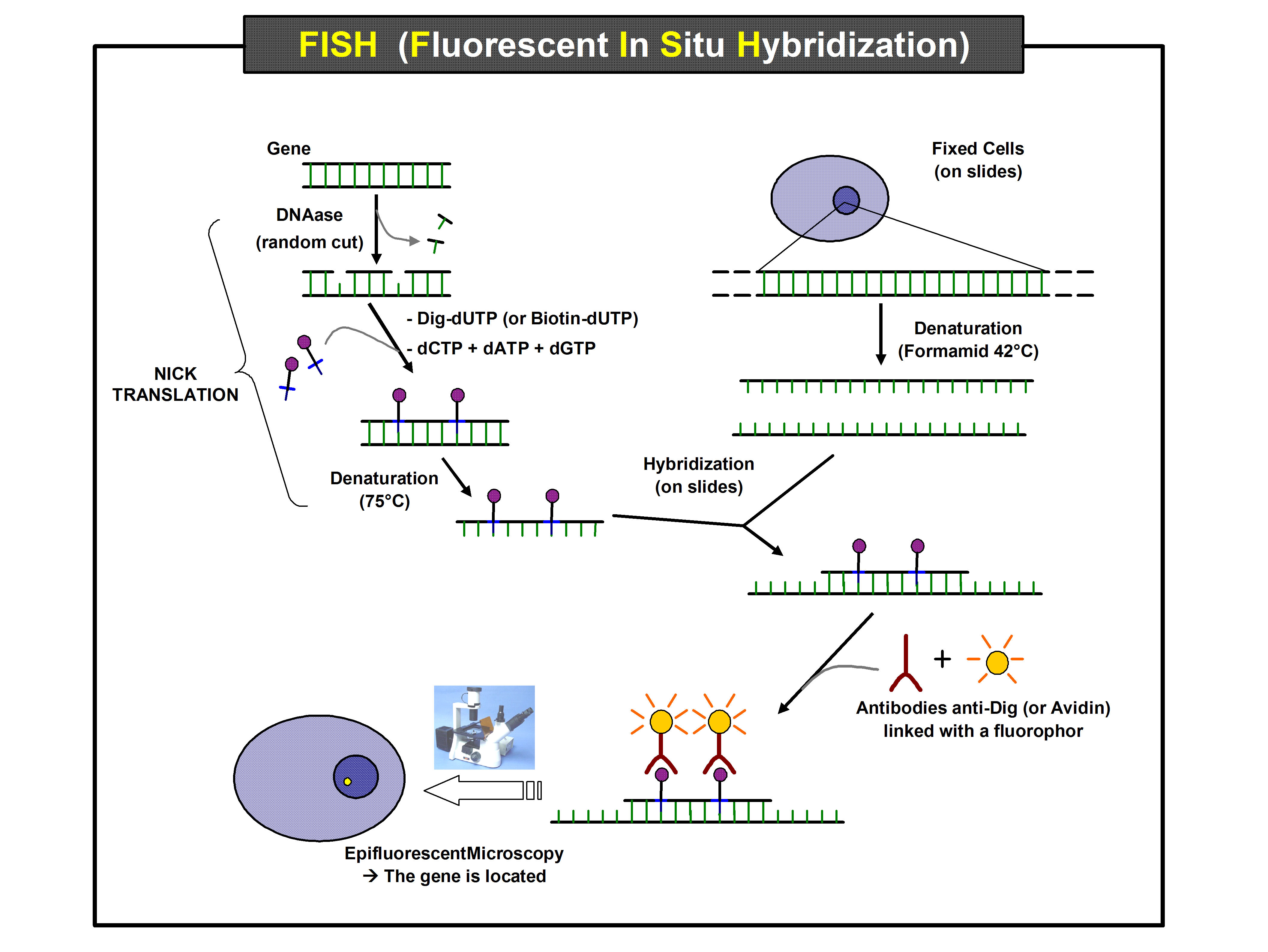 First, a probe is constructed. The probe must be large enough to hybridize specifically with its target but not so large as to impede the hybridization process. The probe is tagged directly with fluorophores, with targets for
First, a probe is constructed. The probe must be large enough to hybridize specifically with its target but not so large as to impede the hybridization process. The probe is tagged directly with fluorophores, with targets for
MA-FISH
uses a microfluidic flow to increase DNA hybridization efficiency, decreasing expensive FISH probe consumption and reduce the hybridization time. MA-FISH is applied for detecting the ''HER2'' gene in breast cancer tissues.
HF-FISH
uses primary additive excitation/emission combination of fluorophores to generate additional spectra through a labeling process known as dynamic optical transmission (DOT). Three primary fluorophores are able to generate a total of 7 readily detectable emission spectra as a result of combinatorial labeling using DOT. Hybrid Fusion FISH enables highly multiplexed FISH applications that are targeted within clinical oncology panels. The technology offers faster scoring with efficient probesets that can be readily detected with traditional fluorescent microscopes.
 FISH can be used to study the evolution of chromosomes. Species that are related have similar chromosomes. This homology can be detected by gene or genome sequencing but also by FISH. For instance, human and
FISH can be used to study the evolution of chromosomes. Species that are related have similar chromosomes. This homology can be detected by gene or genome sequencing but also by FISH. For instance, human and
Image:FISH (technique).gif, Another schematic of FISH process.
Image:FISHchip.jpg, Microfluidic chip that lowered the cost-per-test of FISH by 90%.
Image:BacteriaFISH.jpg, Dual label FISH image; Bifidobacteria Cy3, Total bacteria FITC.
Image:NEAT1 paraspeckles in U-2 OS cells.jpg, Paraspeckles visualized by single-molecule FISH against NEAT1 (Quasar 570) in U-2 OS cells (DAPI).
fiber FISH
from the
guide to fiber FISH
from Octavian Henegariu
Fibre FISH protocol
from the
CARD-FISH, BioMineWiki
Preparation of Complex DNA Probe Sets for 3D FISH with up to Six Different Fluorochromes
Fluorescence in situ Hybridization Photos of bacteria
* Rational design of polynucleotide probe mixes to identify particular genes in defined taxa: www.dnaBaser.com/PolyPro {{DEFAULTSORT:Fluorescent In Situ Hybridization Anatomical pathology Cytogenetics Laboratory techniques Molecular biology Gene tests Pathology Nuclear organization

 Fluorescence ''in situ'' hybridization (FISH) is a molecular cytogenetic technique that uses
Fluorescence ''in situ'' hybridization (FISH) is a molecular cytogenetic technique that uses fluorescent probes
A fluorophore (or fluorochrome, similarly to a chromophore) is a fluorescent chemical compound that can re-emit light upon light excitation. Fluorophores typically contain several combined aromatic groups, or planar or cyclic molecules with sev ...
that bind to only particular parts of a nucleic acid
Nucleic acids are biopolymers, macromolecules, essential to all known forms of life. They are composed of nucleotides, which are the monomers made of three components: a 5-carbon sugar, a phosphate group and a nitrogenous base. The two main cl ...
sequence with a high degree of sequence complementarity. It was developed by biomedical researchers in the early 1980s to detect and localize the presence or absence of specific DNA sequences on chromosomes. Fluorescence microscopy can be used to find out where the fluorescent probe is bound to the chromosomes. FISH is often used for finding specific features in DNA for use in genetic counseling
Genetic counseling is the process of investigating individuals and families affected by or at risk of genetic disorders to help them understand and adapt to the medical, psychological and familial implications of genetic contributions to disease; t ...
, medicine, and species identification. FISH can also be used to detect and localize specific RNA targets ( mRNA, lncRNA and miRNA) in cells, circulating tumor cells, and tissue samples. In this context, it can help define the spatial-temporal patterns of gene expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, protein or non-coding RNA, and ultimately affect a phenotype, as the final effect. The ...
within cells and tissues.
Probes – RNA and DNA
 In biology, a probe is a single strand of DNA or RNA that is complementary to a nucleotide sequence of interest.
RNA probes can be designed for any gene or any sequence within a gene for visualization of mRNA, lncRNA and miRNA in tissues and cells. FISH is used by examining the cellular reproduction cycle, specifically interphase of the nuclei for any chromosomal abnormalities. FISH allows the analysis of a large series of archival cases much easier to identify the pinpointed chromosome by creating a probe with an artificial chromosomal foundation that will attract similar chromosomes. The hybridization signals for each probe when a nucleic abnormality is detected. Each probe for the detection of mRNA and lncRNA is composed of ~20-50 oligonucleotide pairs, each pair covering a space of 40–50 bp. The specifics depend on the specific FISH technique used. For miRNA detection, the probes use proprietary chemistry for specific detection of miRNA and cover the entire miRNA sequence.
In biology, a probe is a single strand of DNA or RNA that is complementary to a nucleotide sequence of interest.
RNA probes can be designed for any gene or any sequence within a gene for visualization of mRNA, lncRNA and miRNA in tissues and cells. FISH is used by examining the cellular reproduction cycle, specifically interphase of the nuclei for any chromosomal abnormalities. FISH allows the analysis of a large series of archival cases much easier to identify the pinpointed chromosome by creating a probe with an artificial chromosomal foundation that will attract similar chromosomes. The hybridization signals for each probe when a nucleic abnormality is detected. Each probe for the detection of mRNA and lncRNA is composed of ~20-50 oligonucleotide pairs, each pair covering a space of 40–50 bp. The specifics depend on the specific FISH technique used. For miRNA detection, the probes use proprietary chemistry for specific detection of miRNA and cover the entire miRNA sequence.
 Probes are often derived from fragments of DNA that were isolated, purified, and amplified for use in the
Probes are often derived from fragments of DNA that were isolated, purified, and amplified for use in the Human Genome Project
The Human Genome Project (HGP) was an international scientific research project with the goal of determining the base pairs that make up human DNA, and of identifying, mapping and sequencing all of the genes of the human genome from both a ...
. The size of the human genome is so large, compared to the length that could be sequenced directly, that it was necessary to divide the genome into fragments. (In the eventual analysis, these fragments were put into order by digesting a copy of each fragment into still smaller fragments using sequence-specific endonucleases, measuring the size of each small fragment using size-exclusion chromatography, and using that information to determine where the large fragments overlapped one another.) To preserve the fragments with their individual DNA sequences, the fragments were added into a system of continually replicating bacteria populations. Clonal populations of bacteria, each population maintaining a single artificial chromosome, are stored in various laboratories around the world. The artificial chromosomes ( BAC) can be grown, extracted, and labeled, in any lab containing a library. Genomic libraries are often named after the institution in which they were developed. An example being the RPCI-11 library, which is named after Roswell Park Comprehensive Cancer Center (formerly known as Roswell Park Cancer Institute) in Buffalo, New York. These fragments are on the order of 100 thousand base-pairs, and are the basis for most FISH probes.
Preparation and hybridization process – RNA
The purpose of using RNA FISH is to detect target mRNA transcripts in cells, tissue sections, or even whole-mounts. The process is done in 3 main procedures: tissue preparation (pre-hybridization), hybridization, and washing (post-hybridization). The tissue preparation starts by collecting the appropriate tissue sections to perform RNA FISH. First, cells,circulating tumor cell A circulating tumor cell (CTC) is a cell that has shed into the vasculature or lymphatics from a primary tumor and is carried around the body in the blood circulation. CTCs can extravasate and become ''seeds'' for the subsequent growth of additional ...
s (CTCs), formalin-fixed paraffin-embedded (FFPE), or frozen tissue sections are fixed. Some commonly used fixatives are 4% formaldehyde or paraformaldehyde (PFA) in phosphate buffered saline (PBS). FISH has also been successfully done on unfixed cells. After fixation, samples are permeabilized to allow the penetration of hybridization reagents. The use of detergents at a 0.1% concentration is commonly used to enhance the tissue permeability such as Tween-20 or Triton X-100.
It is critical for the hybridization process to have all optimal conditions to have a successful in situ result, including temperature, pH, salt concentration, and time of the hybridization reaction. After checking all the necessary conditions, hybridization steps can be started by first adding a target-specific probe, composed of 20 oligonucleotide pairs, hybridizes to the target RNA(s). Separate but compatible signal amplification systems enable the multiplex assay (up to two targets per assay). Signal amplification is achieved via series of sequential hybridization steps.
After the hybridization steps, washing steps are performed. These steps aim to remove nonspecific hybrids and get rid of unbound probe molecules from the samples to reduce any background signaling. The use of ethanol washes are typically used at this stage to reduce autofluorescence in tissues or cells. At the end of the assay the tissue samples are visualized under a fluorescence microscope such as the confocal fluorescence microscope and the Keyence microscope.
Preparation and hybridization process – DNA
 First, a probe is constructed. The probe must be large enough to hybridize specifically with its target but not so large as to impede the hybridization process. The probe is tagged directly with fluorophores, with targets for
First, a probe is constructed. The probe must be large enough to hybridize specifically with its target but not so large as to impede the hybridization process. The probe is tagged directly with fluorophores, with targets for antibodies
An antibody (Ab), also known as an immunoglobulin (Ig), is a large, Y-shaped protein used by the immune system to identify and neutralize foreign objects such as pathogenic bacteria and viruses. The antibody recognizes a unique molecule of the ...
or with biotin
Biotin (or vitamin B7) is one of the B vitamins. It is involved in a wide range of metabolic processes, both in humans and in other organisms, primarily related to the utilization of fats, carbohydrates, and amino acids. The name ''biotin'', bor ...
. Tagging can be done in various ways, such as nick translation, or polymerase chain reaction using tagged nucleotides.
Then, an interphase or metaphase chromosome preparation is produced. The chromosomes are firmly attached to a substrate
Substrate may refer to:
Physical layers
*Substrate (biology), the natural environment in which an organism lives, or the surface or medium on which an organism grows or is attached
** Substrate (locomotion), the surface over which an organism lo ...
, usually glass. Repetitive DNA sequences must be blocked by adding short fragments of DNA to the sample. The probe is then applied to the chromosome DNA and incubated for approximately 12 hours while hybridizing. Several wash steps remove all unhybridized or partially hybridized probes. The results are then visualized and quantified using a microscope that is capable of exciting the dye and recording images.
If the fluorescent signal is weak, amplification of the signal may be necessary in order to exceed the detection threshold of the microscope. Fluorescent signal strength depends on many factors such as probe labeling efficiency, the type of probe, and the type of dye. Fluorescently tagged antibodies
An antibody (Ab), also known as an immunoglobulin (Ig), is a large, Y-shaped protein used by the immune system to identify and neutralize foreign objects such as pathogenic bacteria and viruses. The antibody recognizes a unique molecule of the ...
or streptavidin
Streptavidin is a 66.0 (tetramer) kDa protein purified from the bacterium '' Streptomyces avidinii''. Streptavidin homo-tetramers have an extraordinarily high affinity for biotin (also known as vitamin B7 or vitamin H). With a dissociation co ...
are bound to the dye molecule. These secondary components are selected so that they have a strong signal.
Variations on probes and analysis
FISH is a very general technique. The differences between the various FISH techniques are usually due to variations in the sequence and labeling of the probes; and how they are used in combination. Probes are divided into two generic categories: cellular and acellular. In fluorescent "in situ" hybridization refers to the cellular placement of the probe Probe size is important because shorter probes hybridize less specifically than longer probes, so that long enough strands of DNA or RNA (often 10–25 nucleotides) which are complementary to a given target sequence are often used to locate a target. The overlap defines the resolution of detectable features. For example, if the goal of an experiment is to detect the breakpoint of atranslocation
Translocation may refer to:
* Chromosomal translocation, a chromosome abnormality caused by rearrangement of parts
** Robertsonian translocation, a chromosomal rearrangement in pairs 13, 14, 15, 21, and 22
** Nonreciprocal translocation, transfer ...
, then the overlap of the probes — the degree to which one DNA sequence is contained in the adjacent probes — defines the minimum window in which the breakpoint may be detected.
The mixture of probe sequences determines the type of feature the probe can detect. Probes that hybridize along an entire chromosome are used to count the number of a certain chromosome, show translocations, or identify extra-chromosomal fragments of chromatin. This is often called "whole-chromosome painting." If every possible probe is used, every chromosome, (the whole genome) would be marked fluorescently, which would not be particularly useful for determining features of individual sequences. However, it is possible to create a mixture of smaller probes that are specific to a particular region (locus) of DNA; these mixtures are used to detect deletion mutations. When combined with a specific color, a locus-specific probe mixture is used to detect very specific translocations. Special locus-specific probe mixtures are often used to count chromosomes, by binding to the centromeric regions of chromosomes, which are distinctive enough to identify each chromosome (with the exception of Chromosome 13, 14, 21, 22.)
A variety of other techniques uses mixtures of differently colored probes. A range of colors in mixtures of fluorescent dyes can be detected, so each human chromosome can be identified by a characteristic color using whole-chromosome probe mixtures and a variety of ratios of colors. Although there are more chromosomes than easily distinguishable fluorescent dye colors, ratios of probe mixtures can be used to create ''secondary'' colors. Similar to comparative genomic hybridization, the probe mixture for the secondary colors is created by mixing the correct ratio of two sets of differently colored probes for the same chromosome. This technique is sometimes called M-FISH.
The same physics that make a variety of colors possible for M-FISH can be used for the detection of translocations. That is, colors that are adjacent appear to overlap; a secondary color is observed. Some assays are designed so that the secondary color will be present or absent in cases of interest. An example is the detection of BCR/ABL
The Philadelphia chromosome or Philadelphia translocation (Ph) is a specific genetic abnormality in chromosome 22 of leukemia cancer cells (particularly chronic myeloid leukemia (CML) cells). This chromosome is defective and unusually short becaus ...
translocations, where the secondary color indicates disease. This variation is often called double-fusion FISH or D-FISH. In the opposite situation—where the absence of the secondary color is pathological—is illustrated by an assay used to investigate translocations where only one of the breakpoints is known or constant. Locus-specific probes are made for one side of the breakpoint and the other intact chromosome. In normal cells, the secondary color is observed, but only the primary colors are observed when the translocation occurs. This technique is sometimes called "break-apart FISH".
Single-molecule RNA FISH
Single-molecule RNA FISH, also known as Stellaris® RNA FISH or smFISH, is a method of detecting and quantifying mRNA and other long RNA molecules in a thin layer of tissue sample. Targets can be reliably imaged through the application of multiple short singly labeled oligonucleotide probes. The binding of up to 48 fluorescent labeled oligos to a single molecule of mRNA provides sufficient fluorescence to accurately detect and localize each target mRNA in a wide-fieldfluorescent microscopy
A fluorescence microscope is an optical microscope that uses fluorescence instead of, or in addition to, scattering, reflection, and attenuation or absorption, to study the properties of organic or inorganic substances. "Fluorescence micr ...
image. Probes not binding to the intended sequence do not achieve sufficient localized fluorescence to be distinguished from background.
Single-molecule RNA FISH assays can be performed in simplex or multiplex, and can be used as a follow-up experiment to quantitative PCR, or imaged simultaneously with a fluorescent antibody assay. The technology has potential applications in cancer diagnosis
Cancer is a group of diseases involving Cell growth#Disorders, abnormal cell growth with the potential to Invasion (cancer), invade or Metastasis, spread to other parts of the body. These contrast with benign tumors, which do not spread. Poss ...
, neuroscience, gene expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, protein or non-coding RNA, and ultimately affect a phenotype, as the final effect. The ...
analysis, and companion diagnostics.
Fiber FISH
In an alternative technique to interphase or metaphase preparations, fiber FISH, interphase chromosomes are attached to a slide in such a way that they are stretched out in a straight line, rather than being tightly coiled, as in conventional FISH, or adopting achromosome territory
In cell biology, chromosome territories are regions of the nucleus preferentially occupied by particular chromosomes.
Interphase chromosomes are long DNA strands that are extensively folded, and are often described as appearing like a bowl of s ...
conformation, as in interphase FISH. This is accomplished by applying mechanical shear along the length of the slide, either to cells that have been fixed to the slide and then lysed, or to a solution of purified DNA. A technique known as chromosome combing
Chromosome combing (also known as molecular combing or DNA combing) is a technique used to produce an array of uniformly stretched DNA that is then highly suitable for nucleic acid hybridization studies such as fluorescent in situ hybridisation ...
is increasingly used for this purpose. The extended conformation of the chromosomes allows dramatically higher resolution – even down to a few kilobases. The preparation of fiber FISH samples, although conceptually simple, is a rather skilled art, and only specialized laboratories use the technique routinely.
Q-FISH
Q-FISH combines FISH withPNAs
''Proceedings of the National Academy of Sciences of the United States of America'' (often abbreviated ''PNAS'' or ''PNAS USA'') is a peer-reviewed multidisciplinary scientific journal. It is the official journal of the National Academy of Scien ...
and computer software to quantify fluorescence intensity. This technique is used routinely in telomere length research.
Flow-FISH
Flow-FISH usesflow cytometry
Flow cytometry (FC) is a technique used to detect and measure physical and chemical characteristics of a population of cells or particles.
In this process, a sample containing cells or particles is suspended in a fluid and injected into the flo ...
to perform FISH automatically using per-cell fluorescence measurements.
MA-FISH
Microfluidics-assisted FISHMA-FISH
uses a microfluidic flow to increase DNA hybridization efficiency, decreasing expensive FISH probe consumption and reduce the hybridization time. MA-FISH is applied for detecting the ''HER2'' gene in breast cancer tissues.
MAR-FISH
Microautoradiography FISH is a technique to combine radio-labeled substrates with conventional FISH to detect phylogenetic groups and metabolic activities simultaneously.Hybrid Fusion-FISH
Hybrid Fusion FISHHF-FISH
uses primary additive excitation/emission combination of fluorophores to generate additional spectra through a labeling process known as dynamic optical transmission (DOT). Three primary fluorophores are able to generate a total of 7 readily detectable emission spectra as a result of combinatorial labeling using DOT. Hybrid Fusion FISH enables highly multiplexed FISH applications that are targeted within clinical oncology panels. The technology offers faster scoring with efficient probesets that can be readily detected with traditional fluorescent microscopes.
MERFISH
Multiplexed error-robust fluorescence in situ hybridization is a highly multiplexed version of smFISH. It uses combinatorial labeling, followed by imaging, and then error-resistant encoding to capture a high number of RNA molecules and spatial localization within the cell. The capture of a large number of RNA molecules enables elucidation of gene regulatory networks, prediction of function of unannotated genes, and identification of RNA molecule distribution patterns, which correlate with their associated proteins.STARFISH
Starfish is a set of software tools developed in 2019 by a consortium of scientists to analyze data from nine different variations of FISH, since all variations produce the same set of data—gene expression values mapped to x and y coordinates in a cell. The software, created for all scientists, not just bioinformaticians, reads a set of images, removes noise, and identifies RNA molecules. This approach has set out to define a standard analysis scheme of FISH datasets in a similar way to single-cell transcriptomics analysis.Medical applications
Often parents of children with a developmental disability want to know more about their child's conditions before choosing to have another child. These concerns can be addressed by analysis of the parents' and child's DNA. In cases where the child's developmental disability is not understood, the cause of it can potentially be determined using FISH and cytogenetic techniques. Examples of diseases that are diagnosed using FISH include Prader-Willi syndrome,Angelman syndrome
Angelman syndrome or Angelman's syndrome (AS) is a genetic disorder that mainly affects the nervous system. Symptoms include a small head and a specific facial appearance, severe intellectual disability, developmental disability, limited to no f ...
, 22q13 deletion syndrome
22q13 deletion syndrome, also known as Phelan–McDermid syndrome (PMS), is a genetic disorder caused by deletions or rearrangements on the q terminal end (long arm) of chromosome 22. Any abnormal genetic variation in the q13 region that present ...
, chronic myelogenous leukemia
Chronic myelogenous leukemia (CML), also known as chronic myeloid leukemia, is a cancer of the white blood cells. It is a form of leukemia characterized by the increased and unregulated growth of myeloid cells in the bone marrow and the accumulat ...
, acute lymphoblastic leukemia
Acute lymphoblastic leukemia (ALL) is a cancer of the lymphoid line of blood cells characterized by the development of large numbers of immature lymphocytes. Symptoms may include feeling tired, pale skin color, fever, easy bleeding or bruisin ...
, Cri-du-chat, Velocardiofacial syndrome, and Down syndrome. FISH on sperm cell
Sperm is the male reproductive cell, or gamete, in anisogamous forms of sexual reproduction (forms in which there is a larger, female reproductive cell and a smaller, male one). Animals produce motile sperm with a tail known as a flagellum, whi ...
s is indicated for men with an abnormal somatic or meiotic karyotype
A karyotype is the general appearance of the complete set of metaphase chromosomes in the cells of a species or in an individual organism, mainly including their sizes, numbers, and shapes. Karyotyping is the process by which a karyotype is disce ...
as well as those with oligozoospermia, since approximately 50% of oligozoospermic men have an increased rate of sperm chromosome abnormalities. The analysis of chromosomes 21, X, and Y is enough to identify oligozoospermic individuals at risk.
In medicine, FISH can be used to form a diagnosis, to evaluate prognosis
Prognosis (Greek: πρόγνωσις "fore-knowing, foreseeing") is a medical term for predicting the likely or expected development of a disease, including whether the signs and symptoms will improve or worsen (and how quickly) or remain stabl ...
, or to evaluate remission
Remission often refers to:
*Forgiveness
Remission may also refer to:
Healthcare and science
*Remission (medicine), the state of absence of disease activity in patients with a chronic illness, with the possibility of return of disease activity
*R ...
of a disease, such as cancer. Treatment can then be specifically tailored. A traditional exam involving metaphase chromosome analysis is often unable to identify features that distinguish one disease from another, due to subtle chromosomal features; FISH can elucidate these differences. FISH can also be used to detect diseased cells more easily than standard Cytogenetic methods, which require dividing cells and requires labor and time-intensive manual preparation and analysis of the slides by a technologist. FISH, on the other hand, does not require living cells and can be quantified automatically, a computer counts the fluorescent dots present. However, a trained technologist is required to distinguish subtle differences in banding patterns on bent and twisted metaphase chromosomes. FISH can be incorporated into Lab-on-a-chip microfluidic device. This technology is still in a developmental stage but, like other lab on a chip methods, it may lead to more portable diagnostic techniques.

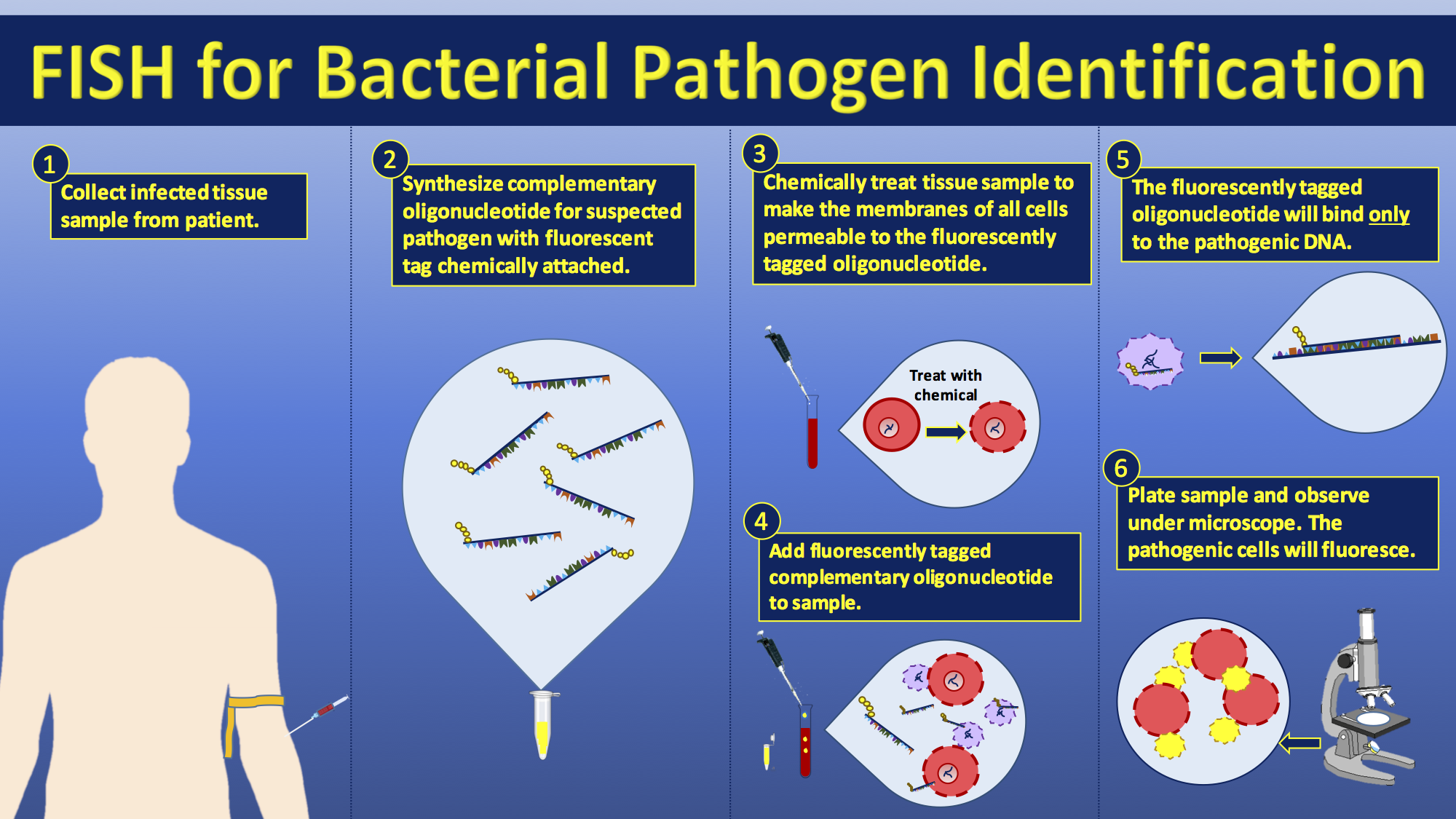
Species identification
FISH has been extensively studied as a diagnostic technique for the identification of pathogens in the field of medical microbiology. Although it has been proven to be a useful and applicable technique, it is still not widely applied in diagnostic laboratories. The short time to diagnosis (less than 2 hours) has been a major advantage compared with biochemical differentiation, but this advantage is challenged by MALDI-TOF-MS which allows the identification of a wider range of pathogens compared with biochemical differentiation techniques. Using FISH for diagnostic purposes has found its purpose when immediate species identification is needed, specifically for the investigation of blood cultures for which FISH is a cheap and easy technique for preliminary rapid diagnosis. FISH can also be used to compare the genomes of two biological species, to deduce evolutionary relationships. A similar hybridization technique is called a zoo blot. Bacterial FISH probes are often primers for the16s rRNA 16S rRNA may refer to:
* 16S ribosomal RNA
16 S ribosomal RNA (or 16 S rRNA) is the RNA component of the 30S subunit of a prokaryotic ribosome ( SSU rRNA). It binds to the Shine-Dalgarno sequence and provides most of the SSU structure.
The g ...
region.
FISH is widely used in the field of microbial ecology, to identify microorganisms
A microorganism, or microbe,, ''mikros'', "small") and ''organism'' from the el, ὀργανισμός, ''organismós'', "organism"). It is usually written as a single word but is sometimes hyphenated (''micro-organism''), especially in olde ...
. Biofilms, for example, are composed of complex (often) multi-species bacterial organizations. Preparing DNA probes for one species and performing FISH with this probe allows one to visualize the distribution of this specific species within the biofilm. Preparing probes (in two different colors) for two species allows researchers to visualize/study co-localization of these two species in the biofilm and can be useful in determining the fine architecture of the biofilm.
Comparative genomic hybridization
Comparative genomic hybridization can be described as a method that uses FISH in a parallel manner with the comparison of the hybridization strength to recall any major disruptions in the duplication process of the DNA sequences in the genome of the nucleus.Virtual karyotype
Virtual karyotyping is another cost-effective, clinically available alternative to FISH panels using thousands to millions of probes on a single array to detect copy number changes, genome-wide, at unprecedented resolution. Currently, this type of analysis will only detect gains and losses of chromosomal material and will not detect balanced rearrangements, such as translocations and inversions which are hallmark aberrations seen in many types of leukemia and lymphoma.Spectral karyotype
Spectral karyotyping is an image of colored chromosomes. Spectral karyotyping involves FISH using multiple forms of many types of probes with the result to see each chromosome labeled through its metaphase stage. This type of karyotyping is used specifically when seeking out chromosome arrangements.Chromosome evolution
 FISH can be used to study the evolution of chromosomes. Species that are related have similar chromosomes. This homology can be detected by gene or genome sequencing but also by FISH. For instance, human and
FISH can be used to study the evolution of chromosomes. Species that are related have similar chromosomes. This homology can be detected by gene or genome sequencing but also by FISH. For instance, human and chimpanzee
The chimpanzee (''Pan troglodytes''), also known as simply the chimp, is a species of great ape native to the forest and savannah of tropical Africa. It has four confirmed subspecies and a fifth proposed subspecies. When its close relative th ...
chromosomes are very similar and FISH can demonstrate that two chimpanzee chromosomes fused to result in one human chromosome. Similarly, species that are more distantly related, have similar chromosomes but with increasing distance chromosomes tend to break and fuse and thus result in mosaic chromosomes. This can be impressively demonstrated by FISH (see figure).
See also
* Chromogenic in situ hybridization (CISH) *Eukaryotic chromosome fine structure
Eukaryotic chromosome fine structure refers to the structure of sequences for eukaryotic chromosomes. Some fine sequences are included in more than one class, so the classification listed is not intended to be completely separate.
Chromosomal char ...
* G banding
* Gene mapping
* Genome evolution
* Happy mapping
* In situ hybridization, the technique used for labelling
* Molecular cytogenetics
* Virtual karyotype
Gallery
References
Further reading
* * *External links
* * Information ofiber FISH
from the
Olympus Corporation
is a Japanese manufacturer of optics and reprography products. Olympus was established on 12 October 1919, initially specializing in microscopes and thermometers. Olympus holds roughly a 70-percent share of the global endoscope market, estimated ...
* guide to fiber FISH
from Octavian Henegariu
Fibre FISH protocol
from the
Human Genome Project
The Human Genome Project (HGP) was an international scientific research project with the goal of determining the base pairs that make up human DNA, and of identifying, mapping and sequencing all of the genes of the human genome from both a ...
at the Sanger Centre
CARD-FISH, BioMineWiki
Preparation of Complex DNA Probe Sets for 3D FISH with up to Six Different Fluorochromes
Fluorescence in situ Hybridization Photos of bacteria
* Rational design of polynucleotide probe mixes to identify particular genes in defined taxa: www.dnaBaser.com/PolyPro {{DEFAULTSORT:Fluorescent In Situ Hybridization Anatomical pathology Cytogenetics Laboratory techniques Molecular biology Gene tests Pathology Nuclear organization