Histone code on:
[Wikipedia]
[Google]
[Amazon]
The histone code is a
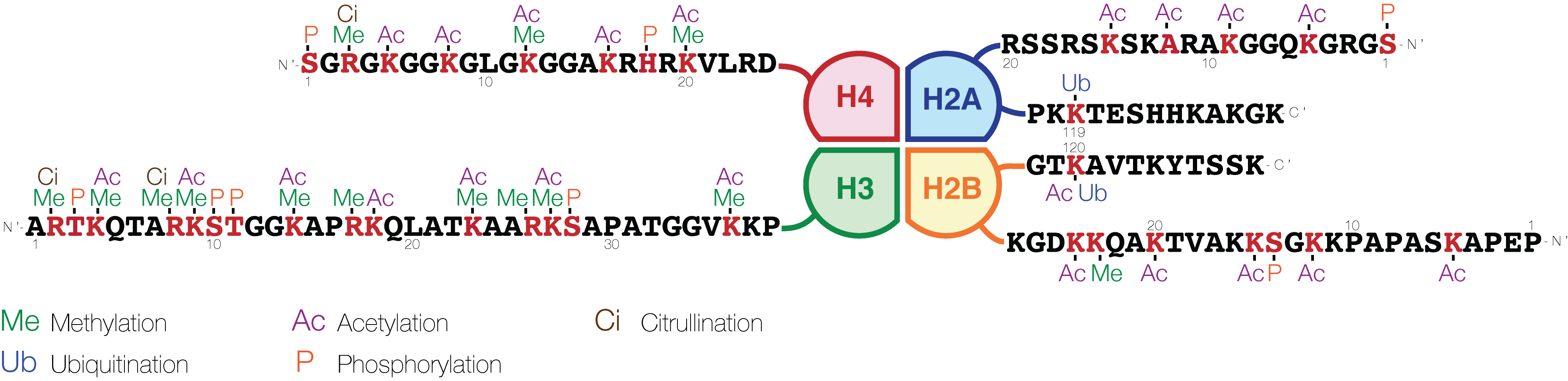
Cellsignal.com Histone Modifications with function and attached references
Histone Code Overview sheet
Histone Modification Guide
{{DEFAULTSORT:Histone Code Epigenetics de:Histonmodifikation#Histon Code
hypothesis
A hypothesis (plural hypotheses) is a proposed explanation for a phenomenon. For a hypothesis to be a scientific hypothesis, the scientific method requires that one can test it. Scientists generally base scientific hypotheses on previous obse ...
that the transcription of genetic information encoded in DNA is in part regulated by chemical modifications (known as ''histone marks'') to histone
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei. They act as spools around which DNA winds to create structural units called nucleosomes. Nucleosomes in turn a ...
proteins, primarily on their unstructured ends. Together with similar modifications such as DNA methylation it is part of the epigenetic code. Histones associate with DNA to form nucleosomes, which themselves bundle to form chromatin
Chromatin is a complex of DNA and protein found in eukaryote, eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important ...
fibers, which in turn make up the more familiar chromosome
A chromosome is a long DNA molecule with part or all of the genetic material of an organism. In most chromosomes the very long thin DNA fibers are coated with packaging proteins; in eukaryotic cells the most important of these proteins ar ...
. Histones are globular proteins with a flexible N-terminus (taken to be the tail) that protrudes from the nucleosome. Many of the histone tail modifications correlate very well to chromatin structure and both histone modification state and chromatin structure correlate well to gene expression levels. The critical concept of the histone code hypothesis is that the histone modifications serve to recruit other proteins by specific recognition of the modified histone via protein domains specialized for such purposes, rather than through simply stabilizing or destabilizing the interaction between histone and the underlying DNA. These recruited proteins then act to alter chromatin structure actively or to promote transcription.
For details of gene expression regulation by histone modifications see table below.
The hypothesis
The hypothesis is thatchromatin
Chromatin is a complex of DNA and protein found in eukaryote, eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important ...
-DNA interactions are guided by combinations of histone modifications. While it is accepted that modifications (such as methylation, acetylation, ADP-ribosylation, ubiquitination, citrullination, SUMO-ylation and phosphorylation
In chemistry, phosphorylation is the attachment of a phosphate group to a molecule or an ion. This process and its inverse, dephosphorylation, are common in biology and could be driven by natural selection. Text was copied from this source, wh ...
) to histone
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei. They act as spools around which DNA winds to create structural units called nucleosomes. Nucleosomes in turn a ...
tails alter chromatin structure, a complete understanding of the precise mechanisms by which these alterations to histone tails influence DNA-histone interactions remains elusive. However, some specific examples have been worked out in detail. For example, phosphorylation of serine residues 10 and 28 on histone H3
Histone H3 is one of the five main histones involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and a long N-terminal tail, H3 is involved with the structure of the nucleosomes of the 'beads on a st ...
is a marker for chromosomal condensation. Similarly, the combination of phosphorylation of serine residue 10 and acetylation of a lysine residue 14 on histone H3 is a tell-tale sign of active transcription.

Modifications
Well characterized modifications to histones include: * Methylation: Both lysine and arginine residues are known to be methylated. Methylated lysines are the best understood marks of the histone code, as specific methylated lysine match well with gene expression states. Methylation of lysines H3K4 and H3K36 is correlated with transcriptional activation while demethylation of H3K4 is correlated with silencing of the genomic region. Methylation of lysines H3K9 and H3K27 is correlated with transcriptional repression. Particularly, H3K9me3 is highly correlated with constitutive heterochromatin. Methylation of histone lysine also has a role inDNA repair
DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as radiation can cause DNA d ...
. For instance, H3K36me3 is required for homologous recombination
Homologous recombination is a type of genetic recombination in which genetic information is exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acids (usually DNA as in cellular organisms but may ...
al repair of DNA double-strand breaks, and H4K20me2 facilitates repair of such breaks by non-homologous end joining
Non-homologous end joining (NHEJ) is a pathway that repairs double-strand breaks in DNA. NHEJ is referred to as "non-homologous" because the break ends are directly ligated without the need for a homologous template, in contrast to homology direc ...
.
* Acetylation—by HAT (histone acetyl transferase); deacetylation—by HDAC
Histone deacetylases (, HDAC) are a class of enzymes that remove acetyl groups (O=C-CH3) from an ε-N-acetyl lysine amino acid on a histone, allowing the histones to wrap the DNA more tightly. This is important because DNA is wrapped around his ...
(histone deacetylase): Acetylation tends to define the 'openness' of chromatin
Chromatin is a complex of DNA and protein found in eukaryote, eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important ...
as acetylated histones cannot pack as well together as deacetylated histones.
*Phosphorylation
In chemistry, phosphorylation is the attachment of a phosphate group to a molecule or an ion. This process and its inverse, dephosphorylation, are common in biology and could be driven by natural selection. Text was copied from this source, wh ...
* Ubiquitination
* SUMOylation
However, there are many more histone modifications, and sensitive mass spectrometry approaches have recently greatly expanded the catalog.
A very basic summary of the histone code for gene expression status is given below (histone nomenclature is described here
Here is an adverb that means "in, on, or at this place". It may also refer to:
Software
* Here Technologies, a mapping company
* Here WeGo (formerly Here Maps), a mobile app and map website by Here
Television
* Here TV (formerly "here!"), a ...
):
Histone H2B
Histone H2B is one of the 5 main histone proteins involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and long N-terminal and C-terminal tails, H2B is involved with the structure of the nucleosomes.
S ...
* H2BK5ac
Histone H3
Histone H3 is one of the five main histones involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and a long N-terminal tail, H3 is involved with the structure of the nucleosomes of the 'beads on a st ...
* H3K4me1 - primed enhancers
* H3K4me3 is enriched in transcriptionally active promoters.
* H3K9me2 -repression
* H3K9me3 is found in constitutively repressed genes.
* H3K27me3 is found in facultatively repressed genes.
* H3K36me
* H3K36me2
* H3K36me3 is found in actively transcribed gene bodies.
* H3K79me2
H3K79me2 is an epigenetic modification to the DNA packaging protein Histone H3. It is a mark that indicates the di-methylation at the 79th lysine residue of the histone H3 protein. H3K79me2 is detected in the transcribed regions of active genes.
...
* H3K9ac is found in actively transcribed promoters.
* H3K14ac is found in actively transcribed promoters.
* H3K23ac
* H3K27ac distinguishes active enhancers from poised enhancers.
* H3K36ac
* H3K56ac is a proxy for de novo histone assembly.
* H3K122ac is enriched in poised promoters and also found in a different type of putative enhancer that lacks H3K27ac.
Histone H4
Histone H4 is one of the five main histone proteins involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and a long N-terminal tail, H4 is involved with the structure of the nucleosome of the 'beads on ...
* H4K5ac
H4K5ac is an epigenetic modification to the DNA packaging protein histone H4. It is a mark that indicates the acetylation at the 5th lysine residue of the histone H4 protein. H4K5 is the closest lysine residue to the N-terminal tail of histone H4. ...
* H4K8ac
H4K8ac, representing an epigenetic modification to the DNA packaging protein histone H4, is a mark indicating the acetylation at the 8th lysine residue of the histone H4 protein. It has been implicated in the prevalence of malaria.
Nomenclature
...
* H4K12ac
H4K12ac is an epigenetic modification to the DNA packaging protein histone H4. It is a mark that indicates the acetylation at the 12th lysine residue of the histone H4 protein. H4K12ac is involved in learning and memory. It is possible that resto ...
* H4K16ac
H4K16ac is an epigenetic modification to the DNA packaging protein Histone H4. It is a mark that indicates the acetylation at the 16th lysine residue of the histone H4 protein.
H4K16ac is unusual in that it has both transcriptional activation ...
* H4K20me
H4K20me is an epigenetic modification to the DNA packaging protein Histone H4. It is a mark that indicates the mono- methylation at the 20th lysine residue of the histone H4 protein. This mark can be di- and tri-methylated. It is critical for ge ...
* H4K91ac
H4K91ac is an epigenetic modification to the DNA packaging protein histone H4. It is a mark that indicates the acetylation at the 91st lysine residue of the histone H4 protein. No known diseases are attributed to this mark but it might be implicat ...
Complexity
Unlike this simplified model, any real histone code has the potential to be massively complex; each of the four standard histones can be simultaneously modified at multiple different sites with multiple different modifications. To give an idea of this complexity,histone H3
Histone H3 is one of the five main histones involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and a long N-terminal tail, H3 is involved with the structure of the nucleosomes of the 'beads on a st ...
contains nineteen lysines known to be methylated—each can be un-, mono-, di- or tri-methylated. If modifications are independent, this allows a potential 419 or 280 billion different lysine methylation patterns, far more than the maximum number of histones in a human genome (6.4 Gb / ~150 bp = ~44 million histones if they are very tightly packed). And this does not include lysine acetylation (known for H3 at nine residues), arginine methylation (known for H3 at three residues) or threonine/serine/tyrosine phosphorylation (known for H3 at eight residues), not to mention modifications of other histones.
Every nucleosome in a cell can therefore have a different set of modifications, raising the question of whether common patterns of histone modifications exist. A study of about 40 histone modifications across human gene promoters found over 4000 different combinations used, over 3000 occurring at only a single promoter. However, patterns were discovered including a set of 17 histone modifications that are present together at over 3000 genes. Therefore, patterns of histone modifications do occur but they are very intricate, and we currently have detailed biochemical understanding of the importance of a relatively small number of modifications.
Structural determinants of histone recognition by readers, writers and erasers of the histone code are revealed by a growing body of experimental data.
See also
*Histone
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei. They act as spools around which DNA winds to create structural units called nucleosomes. Nucleosomes in turn a ...
* Histone-modifying enzymes
References
External links
Cellsignal.com Histone Modifications with function and attached references
Histone Code Overview sheet
Histone Modification Guide
{{DEFAULTSORT:Histone Code Epigenetics de:Histonmodifikation#Histon Code