Functional Element on:
[Wikipedia]
[Google]
[Amazon]
Functional genomics is a field of 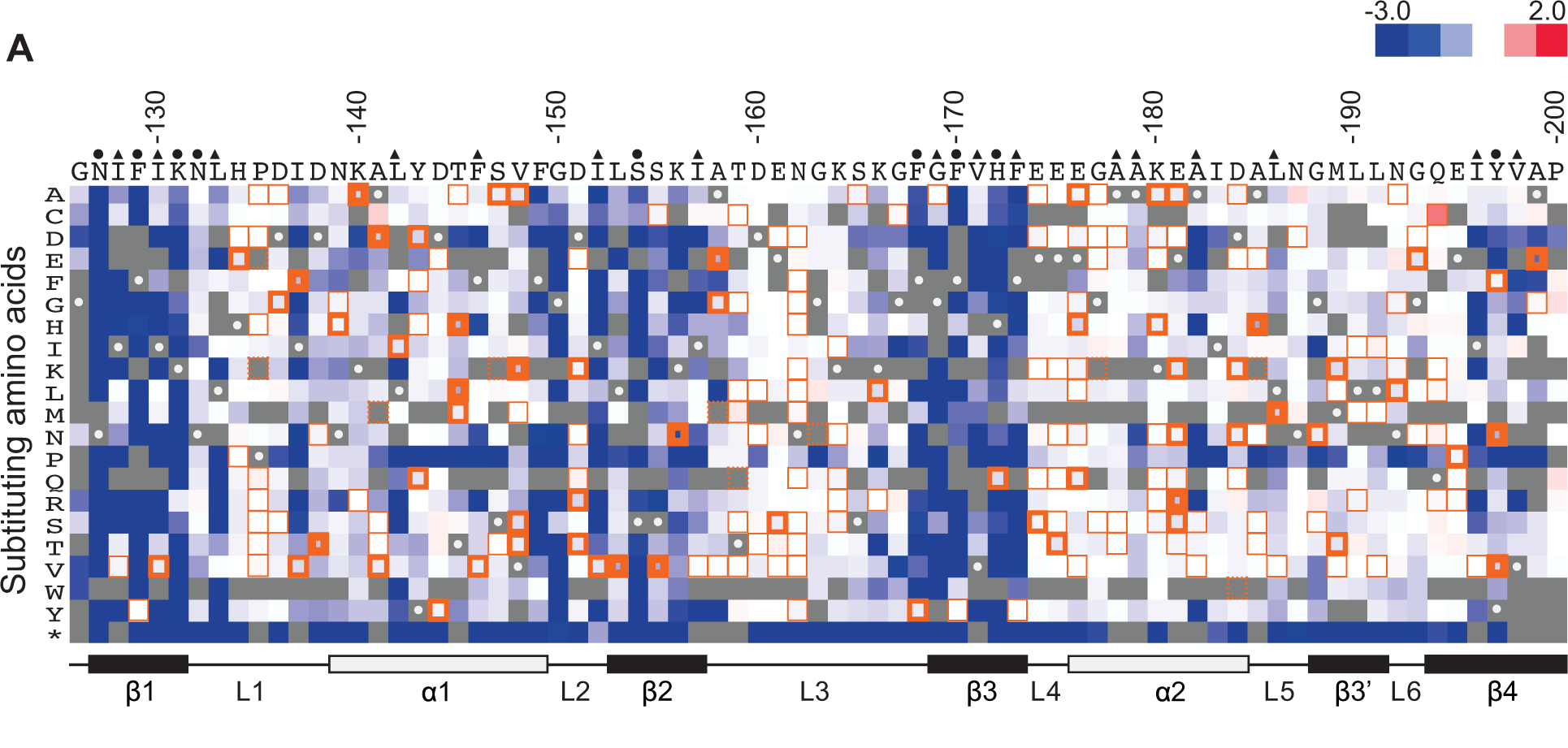
 Microarrays measure the amount of mRNA in a sample that corresponds to a given gene or probe DNA sequence. Probe sequences are immobilized on a solid surface and allowed to
Microarrays measure the amount of mRNA in a sample that corresponds to a given gene or probe DNA sequence. Probe sequences are immobilized on a solid surface and allowed to
 Perturb-seq couples CRISPR mediated gene knockdowns with single-cell gene expression. Linear models are used to calculate the effect of the knockdown of a single gene on the expression of multiple genes.
Perturb-seq couples CRISPR mediated gene knockdowns with single-cell gene expression. Linear models are used to calculate the effect of the knockdown of a single gene on the expression of multiple genes.
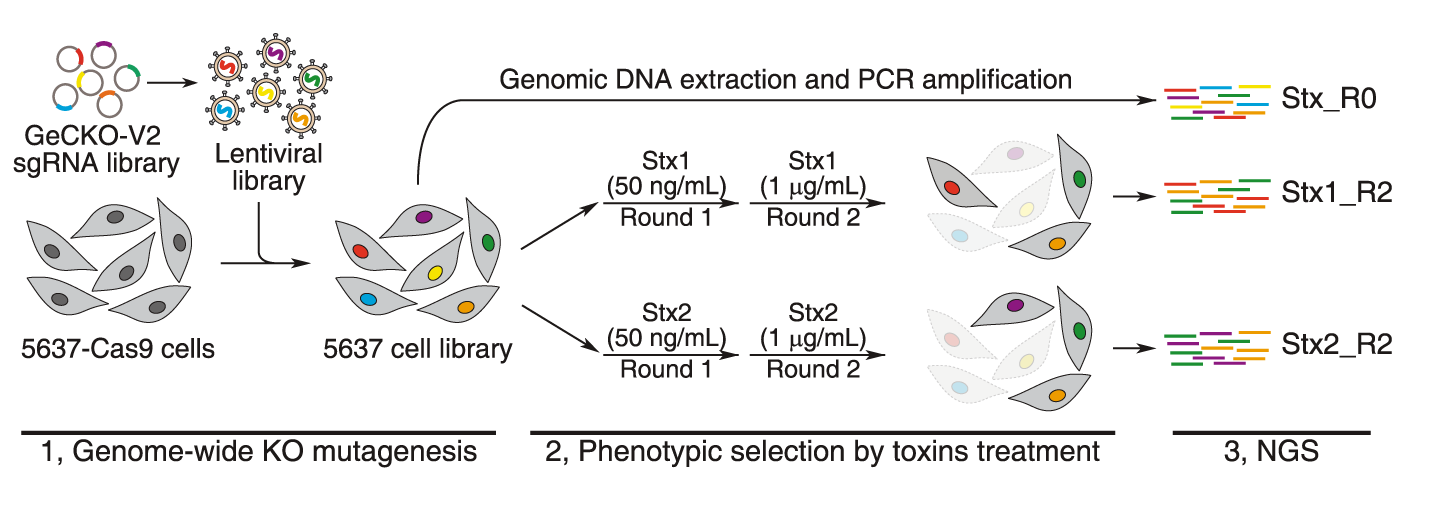 CRISPR-Cas9 has been used to delete genes in a multiplexed manner in cell-lines. Quantifying the amount of guide-RNAs for each gene before and after the experiment can point towards essential genes. If a guide-RNA disrupts an essential gene it will lead to the loss of that cell and hence there will be a depletion of that particular guide-RNA after the screen. In a recent CRISPR-cas9 experiment in mammalian cell-lines, around 2000 genes were found to be essential in multiple cell-lines. Some of these genes were essential in only one cell-line. Most of genes are part of multi-protein complexes. This approach can be used to identify synthetic lethality by using the appropriate genetic background. CRISPRi and CRISPRa enable loss-of-function and gain-of-function screens in a similar manner. CRISPRi identified ~2100 essential genes in the K562 cell-line. CRISPR deletion screens have also been used to identify potential regulatory elements of a gene. For example, a technique called ScanDel was published which attempted this approach. The authors deleted regions outside a gene of interest(HPRT1 involved in a Mendelian disorder) in an attempt to identify regulatory elements of this gene. Gassperini et al. did not identify any distal regulatory elements for HPRT1 using this approach, however such approaches can be extended to other genes of interest.
CRISPR-Cas9 has been used to delete genes in a multiplexed manner in cell-lines. Quantifying the amount of guide-RNAs for each gene before and after the experiment can point towards essential genes. If a guide-RNA disrupts an essential gene it will lead to the loss of that cell and hence there will be a depletion of that particular guide-RNA after the screen. In a recent CRISPR-cas9 experiment in mammalian cell-lines, around 2000 genes were found to be essential in multiple cell-lines. Some of these genes were essential in only one cell-line. Most of genes are part of multi-protein complexes. This approach can be used to identify synthetic lethality by using the appropriate genetic background. CRISPRi and CRISPRa enable loss-of-function and gain-of-function screens in a similar manner. CRISPRi identified ~2100 essential genes in the K562 cell-line. CRISPR deletion screens have also been used to identify potential regulatory elements of a gene. For example, a technique called ScanDel was published which attempted this approach. The authors deleted regions outside a gene of interest(HPRT1 involved in a Mendelian disorder) in an attempt to identify regulatory elements of this gene. Gassperini et al. did not identify any distal regulatory elements for HPRT1 using this approach, however such approaches can be extended to other genes of interest.
 New computational methods have been developed for understanding the results of a deep mutational scanning experiment. 'phydms' compares the result of a deep mutational scanning experiment to a phylogenetic tree. This allows the user to infer if the selection process in nature applies similar constraints on a protein as the results of the deep mutational scan indicate. This may allow an experimenter to choose between different experimental conditions based on how well they reflect nature. Deep mutational scanning has also been used to infer protein-protein interactions. The authors used a thermodynamic model to predict the effects of mutations in different parts of a dimer. Deep mutational structure can also be used to infer protein structure. Strong positive epistasis between two mutations in a deep mutational scan can be indicative of two parts of the protein that are close to each other in 3-D space. This information can then be used to infer protein structure. A proof of principle of this approach was shown by two groups using the protein GB1.
Results from MPRA experiments have required machine learning approaches to interpret the data. A gapped k-mer SVM model has been used to infer the kmers that are enriched within cis-regulatory sequences with high activity compared to sequences with lower activity. These models provide high predictive power. Deep learning and random forest approaches have also been used to interpret the results of these high-dimensional experiments. These models are beginning to help develop a better understanding of non-coding DNA function towards gene-regulation.
New computational methods have been developed for understanding the results of a deep mutational scanning experiment. 'phydms' compares the result of a deep mutational scanning experiment to a phylogenetic tree. This allows the user to infer if the selection process in nature applies similar constraints on a protein as the results of the deep mutational scan indicate. This may allow an experimenter to choose between different experimental conditions based on how well they reflect nature. Deep mutational scanning has also been used to infer protein-protein interactions. The authors used a thermodynamic model to predict the effects of mutations in different parts of a dimer. Deep mutational structure can also be used to infer protein structure. Strong positive epistasis between two mutations in a deep mutational scan can be indicative of two parts of the protein that are close to each other in 3-D space. This information can then be used to infer protein structure. A proof of principle of this approach was shown by two groups using the protein GB1.
Results from MPRA experiments have required machine learning approaches to interpret the data. A gapped k-mer SVM model has been used to infer the kmers that are enriched within cis-regulatory sequences with high activity compared to sequences with lower activity. These models provide high predictive power. Deep learning and random forest approaches have also been used to interpret the results of these high-dimensional experiments. These models are beginning to help develop a better understanding of non-coding DNA function towards gene-regulation.
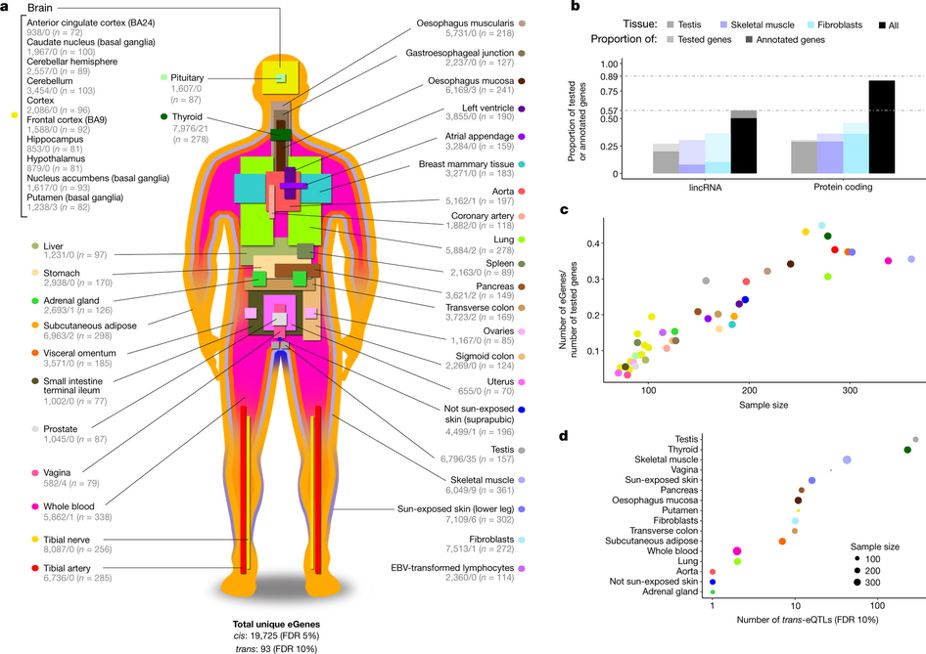 The GTEx project is a human genetics project aimed at understanding the role of genetic variation in shaping variation in the transcriptome across tissues. The project has collected a variety of tissue samples (> 50 different tissues) from more than 700 post-mortem donors. This has resulted in the collection of >11,000 samples. GTEx has helped understand the tissue-sharing and tissue-specificity of
The GTEx project is a human genetics project aimed at understanding the role of genetic variation in shaping variation in the transcriptome across tissues. The project has collected a variety of tissue samples (> 50 different tissues) from more than 700 post-mortem donors. This has resulted in the collection of >11,000 samples. GTEx has helped understand the tissue-sharing and tissue-specificity of
European Science Foundation Programme on Frontiers of Functional GenomicsMUGEN NoE
— Integrated Functional Genomics in Mutant Mouse Models
Nature insights: functional genomicsENCODE
{{DEFAULTSORT:Functional Genomics Molecular biology Genomics
molecular biology
Molecular biology is the branch of biology that seeks to understand the molecular basis of biological activity in and between cells, including biomolecular synthesis, modification, mechanisms, and interactions. The study of chemical and phys ...
that attempts to describe gene
In biology, the word gene (from , ; "... Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a b ...
(and protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, respon ...
) functions and interactions. Functional genomics make use of the vast data generated by genomic
Genomics is an interdisciplinary field of biology focusing on the structure, function, evolution, mapping, and editing of genomes. A genome is an organism's complete set of DNA, including all of its genes as well as its hierarchical, three-dim ...
and transcriptomic projects (such as genome sequencing projects and RNA sequencing). Functional genomics focuses on the dynamic aspects such as gene transcription, translation
Translation is the communication of the Meaning (linguistic), meaning of a #Source and target languages, source-language text by means of an Dynamic and formal equivalence, equivalent #Source and target languages, target-language text. The ...
, regulation of gene expression
Regulation of gene expression, or gene regulation, includes a wide range of mechanisms that are used by cells to increase or decrease the production of specific gene products (protein or RNA). Sophisticated programs of gene expression are wid ...
and protein–protein interaction
Protein–protein interactions (PPIs) are physical contacts of high specificity established between two or more protein molecules as a result of biochemical events steered by interactions that include electrostatic forces, hydrogen bonding and th ...
s, as opposed to the static aspects of the genomic information such as DNA sequence
DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, guanine, cytosine, and thymine. Th ...
or structures. A key characteristic of functional genomics studies is their genome-wide approach to these questions, generally involving high-throughput methods rather than a more traditional "gene-by-gene" approach.

Definition and goals of functional genomics
In order to understand functional genomics it is important to first define function. In their paper Graur et al. define function in two possible ways. These are "selected effect" and "causal role". The "selected effect" function refers to the function for which a trait (DNA, RNA, protein etc.) is selected for. The "causal role" function refers to the function that a trait is sufficient and necessary for. Functional genomics usually tests the "causal role" definition of function. The goal of functional genomics is to understand the function of genes or proteins, eventually all components of a genome. The term functional genomics is often used to refer to the many technical approaches to study an organism's genes and proteins, including the "biochemical, cellular, and/or physiological properties of each and every gene product" while some authors include the study of nongenic elements in their definition. Functional genomics may also include studies of natural genetic variation over time (such as an organism's development) or space (such as its body regions), as well as functional disruptions such as mutations. The promise of functional genomics is to generate and synthesize genomic and proteomic knowledge into an understanding of the dynamic properties of an organism. This could potentially provide a more complete picture of how the genome specifies function compared to studies of single genes. Integration of functional genomics data is often a part ofsystems biology
Systems biology is the computational modeling, computational and mathematical analysis and modeling of complex biological systems. It is a biology-based interdisciplinary field of study that focuses on complex interactions within biological syst ...
approaches.
Techniques and applications
Functional genomics includes function-related aspects of the genome itself such asmutation
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, m ...
and polymorphism
Polymorphism, polymorphic, polymorph, polymorphous, or polymorphy may refer to:
Computing
* Polymorphism (computer science), the ability in programming to present the same programming interface for differing underlying forms
* Ad hoc polymorphis ...
(such as single nucleotide polymorphism (SNP) analysis), as well as the measurement of molecular activities. The latter comprise a number of "-omics
The branches of science known informally as omics are various disciplines in biology whose names end in the suffix ''-omics'', such as genomics, proteomics, metabolomics, metagenomics, phenomics and transcriptomics. Omics aims at the coll ...
" such as transcriptomics
Transcriptomics technologies are the techniques used to study an organism's transcriptome, the sum of all of its RNA transcripts. The information content of an organism is recorded in the DNA of its genome and expressed through transcription. H ...
(gene expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, protein or non-coding RNA, and ultimately affect a phenotype, as the final effect. ...
), proteomics
Proteomics is the large-scale study of proteins. Proteins are vital parts of living organisms, with many functions such as the formation of structural fibers of muscle tissue, enzymatic digestion of food, or synthesis and replication of DNA. In ...
(protein production
Protein production is the biotechnological process of generating a specific protein. It is typically achieved by the manipulation of gene expression in an organism such that it expresses large amounts of a recombinant gene. This includes the ...
), and metabolomics
Metabolomics is the scientific study of chemical processes involving metabolites, the small molecule substrates, intermediates, and products of cell metabolism. Specifically, metabolomics is the "systematic study of the unique chemical fingerprin ...
. Functional genomics uses mostly multiplex techniques to measure the abundance of many or all gene products such as mRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
mRNA is created during the ...
s or protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, respon ...
s within a biological sample. A more focused functional genomics approach might test the function of all variants of one gene and quantify the effects of mutants by using sequencing as a readout of activity. Together these measurement modalities endeavor to quantitate the various biological processes and improve our understanding of gene and protein functions and interactions.
At the DNA level
Genetic interaction mapping
Systematic pairwise deletion of genes or inhibition of gene expression can be used to identify genes with related function, even if they do not interact physically.Epistasis
Epistasis is a phenomenon in genetics in which the effect of a gene mutation is dependent on the presence or absence of mutations in one or more other genes, respectively termed modifier genes. In other words, the effect of the mutation is dep ...
refers to the fact that effects for two different gene knockouts may not be additive; that is, the phenotype that results when two genes are inhibited may be different from the sum of the effects of single knockouts.
DNA/Protein interactions
Proteins formed by the translation of the mRNA (messenger RNA, a coded information from DNA for protein synthesis) play a major role in regulating gene expression. To understand how they regulate gene expression it is necessary to identify DNA sequences that they interact with. Techniques have been developed to identify sites of DNA-protein interactions. These includeChIP-sequencing
ChIP-sequencing, also known as ChIP-seq, is a method used to analyze protein interactions with DNA. ChIP-seq combines chromatin immunoprecipitation (ChIP) with massively parallel DNA sequencing to identify the binding sites of DNA-associated pro ...
, CUT&RUN sequencing
CUT&RUN sequencing, also known as cleavage under targets and release using nuclease, is a method used to analyze protein interactions with DNA. CUT&RUN sequencing combines antibody-targeted controlled cleavage by micrococcal nuclease with massivel ...
and Calling Cards.
DNA accessibility assays
Assays have been developed to identify regions of the genome that are accessible. These regions of open chromatin are candidate regulatory regions. These assays include ATAC-seq, DNase-Seq and FAIRE-Seq.At the RNA level
Microarrays
hybridize
Hybridization (or hybridisation) may refer to:
* Hybridization (biology), the process of combining different varieties of organisms to create a hybrid
* Orbital hybridization, in chemistry, the mixing of atomic orbitals into new hybrid orbitals
* ...
with fluorescently labeled "target" mRNA. The intensity of fluorescence of a spot is proportional to the amount of target sequence that has hybridized to that spot and therefore to the abundance of that mRNA sequence in the sample. Microarrays allow for the identification of candidate genes involved in a given process based on variation between transcript levels for different conditions and shared expression patterns with genes of known function.
SAGE
Serial analysis of gene expression
Serial Analysis of Gene Expression (SAGE) is a transcriptomic technique used by molecular biologists to produce a snapshot of the messenger RNA population in a sample of interest in the form of small tags that correspond to fragments of those tr ...
(SAGE) is an alternate method of analysis based on RNA sequencing rather than hybridization. SAGE relies on the sequencing of 10–17 base pair tags which are unique to each gene. These tags are produced from poly-A mRNA and ligated end-to-end before sequencing. SAGE gives an unbiased measurement of the number of transcripts per cell, since it does not depend on prior knowledge of what transcripts to study (as microarrays do).
RNA sequencing
RNA sequencing has taken over microarray and SAGE technology in recent years, as noted in 2016, and has become the most efficient way to study transcription and gene expression. This is typically done bynext-generation sequencing Massive parallel sequencing or massively parallel sequencing is any of several high-throughput approaches to DNA sequencing using the concept of massively parallel processing; it is also called next-generation sequencing (NGS) or second-generation s ...
.
A subset of sequenced RNAs are small RNAs, a class of non-coding RNA molecules that are key regulators of transcriptional and post-transcriptional gene silencing, or RNA silencing
RNA silencing or RNA interference refers to a family of gene silencing effects by which gene expression is negatively regulated by non-coding RNAs such as microRNAs. RNA silencing may also be defined as sequence-specific regulation of gene expres ...
. Next-generation sequencing is the gold standard tool for non-coding RNA
A non-coding RNA (ncRNA) is a functional RNA molecule that is not translated into a protein. The DNA sequence from which a functional non-coding RNA is transcribed is often called an RNA gene. Abundant and functionally important types of non- ...
discovery, profiling and expression analysis.
Massively Parallel Reporter Assays (MPRAs)
Massively parallel reporter assays is a technology to test the cis-regulatory activity of DNA sequences. MPRAs use aplasmid
A plasmid is a small, extrachromosomal DNA molecule within a cell that is physically separated from chromosomal DNA and can replicate independently. They are most commonly found as small circular, double-stranded DNA molecules in bacteria; howev ...
with a synthetic cis-regulatory element upstream of a promoter driving a synthetic gene such as Green Fluorescent Protein. A library of cis-regulatory elements is usually tested using MPRAs, a library can contain from hundreds to thousands of cis-regulatory elements. The cis-regulatory activity of the elements is assayed by using the downstream reporter activity. The activity of all the library members is assayed in parallel using barcodes for each cis-regulatory element. One limitation of MPRAs is that the activity is assayed on a plasmid and may not capture all aspects of gene regulation observed in the genome.
STARR-seq
STARR-seq is a technique similar to MPRAs to assay enhancer activity of randomly sheared genomic fragments. In the original publication, randomly sheared fragments of the ''Drosophila'' genome were placed downstream of a minimal promoter. Candidate enhancers amongst the randomly sheared fragments will transcribe themselves using the minimal promoter. By using sequencing as a readout and controlling for input amounts of each sequence the strength of putative enhancers are assayed by this method.Perturb-seq
 Perturb-seq couples CRISPR mediated gene knockdowns with single-cell gene expression. Linear models are used to calculate the effect of the knockdown of a single gene on the expression of multiple genes.
Perturb-seq couples CRISPR mediated gene knockdowns with single-cell gene expression. Linear models are used to calculate the effect of the knockdown of a single gene on the expression of multiple genes.
At the protein level
Yeast two-hybrid system
A yeasttwo-hybrid screening
Two-hybrid screening (originally known as yeast two-hybrid system or Y2H) is a molecular biology technique used to discover protein–protein interactions (PPIs) and protein–DNA interactions by testing for physical interactions (such as bind ...
(Y2H) tests a "bait" protein against many potential interacting proteins ("prey") to identify physical protein–protein interactions. This system is based on a transcription factor, originally GAL4, whose separate DNA-binding and transcription activation domains are both required in order for the protein to cause transcription of a reporter gene. In a Y2H screen, the "bait" protein is fused to the binding domain of GAL4, and a library of potential "prey" (interacting) proteins is recombinantly expressed in a vector with the activation domain. In vivo interaction of bait and prey proteins in a yeast cell brings the activation and binding domains of GAL4 close enough together to result in expression of a reporter gene
In molecular biology, a reporter gene (often simply reporter) is a gene that researchers attach to a regulatory sequence of another gene of interest in bacteria, cell culture, animals or plants. Such genes are called reporters because the char ...
. It is also possible to systematically test a library of bait proteins against a library of prey proteins to identify all possible interactions in a cell.
AP/MS
Affinity purification andmass spectrometry
Mass spectrometry (MS) is an analytical technique that is used to measure the mass-to-charge ratio of ions. The results are presented as a '' mass spectrum'', a plot of intensity as a function of the mass-to-charge ratio. Mass spectrometry is u ...
(AP/MS) is able to identify proteins that interact with one another in complexes. Complexes of proteins are allowed to form around a particular "bait" protein. The bait protein is identified using an antibody or a recombinant tag which allows it to be extracted along with any proteins that have formed a complex with it. The proteins are then digested into short peptide
Peptides (, ) are short chains of amino acids linked by peptide bonds. Long chains of amino acids are called proteins. Chains of fewer than twenty amino acids are called oligopeptides, and include dipeptides, tripeptides, and tetrapeptides. ...
fragments and mass spectrometry is used to identify the proteins based on the mass-to-charge ratios of those fragments.
Deep mutational scanning
In deep mutational scanning every possible amino acid change in a given protein is first synthesized. The activity of each of these protein variants is assayed in parallel using barcodes for each variant. By comparing the activity to the wild-type protein, the effect of each mutation is identified. While it is possible to assay every possible single amino-acid change due to combinatorics two or more concurrent mutations are hard to test. Deep mutational scanning experiments have also been used to infer protein structure and protein-protein interactions.Mutagenesis and phenotyping
An important functional feature of genes is the phenotype caused by mutations. Mutants can be produced by random mutations or by directed mutagenesis, including site-directed mutagenesis, deleting complete genes, or other techniques.Knock-outs (gene deletions)
Gene function can be investigated by systematically "knocking out" genes one by one. This is done by either deletion or disruption of function (such as byinsertional mutagenesis In molecular biology, insertional mutagenesis is the creation of mutations of DNA by addition of one or more base pairs. Such insertional mutations can occur naturally, mediated by viruses or transposons, or can be artificially created for resear ...
) and the resulting organisms are screened for phenotypes that provide clues to the function of the disrupted gene. Knock-outs have been produced for whole genomes, i.e. by deleting all genes in a genome. For essential genes
Essential genes are indispensable genes for organisms to grow and reproduce offspring under certain environment. However, being ''essential'' is highly dependent on the circumstances in which an organism lives. For instance, a gene required to dige ...
, this is not possible, so other techniques are used, e.g. deleting a gene while expressing the gene from a plasmid
A plasmid is a small, extrachromosomal DNA molecule within a cell that is physically separated from chromosomal DNA and can replicate independently. They are most commonly found as small circular, double-stranded DNA molecules in bacteria; howev ...
, using an inducible promoter, so that the level of gene product can be changed at will (and thus a "functional" deletion achieved).
Site-directed mutagenesis
Site-directed mutagenesis
Site-directed mutagenesis is a molecular biology method that is used to make specific and intentional mutating changes to the DNA sequence of a gene and any gene products. Also called site-specific mutagenesis or oligonucleotide-directed mutagenesi ...
is used to mutate specific bases (and thus amino acids). This is critical to investigate the function of specific amino acids in a protein, e.g. in the active site of an enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different molecule ...
.
RNAi
RNA interference (RNAi) methods can be used to transiently silence or knockdown gene expression using ~20 base-pair double-stranded RNA typically delivered by transfection of synthetic ~20-mer short-interfering RNA molecules (siRNAs) or by virally encoded short-hairpin RNAs (shRNAs). RNAi screens, typically performed in cell culture-based assays or experimental organisms (such as ''C. elegans'') can be used to systematically disrupt nearly every gene in a genome or subsets of genes (sub-genomes); possible functions of disrupted genes can be assigned based on observedphenotype
In genetics, the phenotype () is the set of observable characteristics or traits of an organism. The term covers the organism's morphology or physical form and structure, its developmental processes, its biochemical and physiological prop ...
s.
CRISPR screens
 CRISPR-Cas9 has been used to delete genes in a multiplexed manner in cell-lines. Quantifying the amount of guide-RNAs for each gene before and after the experiment can point towards essential genes. If a guide-RNA disrupts an essential gene it will lead to the loss of that cell and hence there will be a depletion of that particular guide-RNA after the screen. In a recent CRISPR-cas9 experiment in mammalian cell-lines, around 2000 genes were found to be essential in multiple cell-lines. Some of these genes were essential in only one cell-line. Most of genes are part of multi-protein complexes. This approach can be used to identify synthetic lethality by using the appropriate genetic background. CRISPRi and CRISPRa enable loss-of-function and gain-of-function screens in a similar manner. CRISPRi identified ~2100 essential genes in the K562 cell-line. CRISPR deletion screens have also been used to identify potential regulatory elements of a gene. For example, a technique called ScanDel was published which attempted this approach. The authors deleted regions outside a gene of interest(HPRT1 involved in a Mendelian disorder) in an attempt to identify regulatory elements of this gene. Gassperini et al. did not identify any distal regulatory elements for HPRT1 using this approach, however such approaches can be extended to other genes of interest.
CRISPR-Cas9 has been used to delete genes in a multiplexed manner in cell-lines. Quantifying the amount of guide-RNAs for each gene before and after the experiment can point towards essential genes. If a guide-RNA disrupts an essential gene it will lead to the loss of that cell and hence there will be a depletion of that particular guide-RNA after the screen. In a recent CRISPR-cas9 experiment in mammalian cell-lines, around 2000 genes were found to be essential in multiple cell-lines. Some of these genes were essential in only one cell-line. Most of genes are part of multi-protein complexes. This approach can be used to identify synthetic lethality by using the appropriate genetic background. CRISPRi and CRISPRa enable loss-of-function and gain-of-function screens in a similar manner. CRISPRi identified ~2100 essential genes in the K562 cell-line. CRISPR deletion screens have also been used to identify potential regulatory elements of a gene. For example, a technique called ScanDel was published which attempted this approach. The authors deleted regions outside a gene of interest(HPRT1 involved in a Mendelian disorder) in an attempt to identify regulatory elements of this gene. Gassperini et al. did not identify any distal regulatory elements for HPRT1 using this approach, however such approaches can be extended to other genes of interest.
Functional annotations for genes
Genome annotation
Putative genes can be identified by scanning a genome for regions likely to encode proteins, based on characteristics such as longopen reading frames
In molecular biology, open reading frames (ORFs) are defined as spans of DNA sequence between the start and stop codons. Usually, this is considered within a studied region of a prokaryotic DNA sequence, where only one of the six possible readin ...
, transcriptional initiation sequences, and polyadenylation
Polyadenylation is the addition of a poly(A) tail to an RNA transcript, typically a messenger RNA (mRNA). The poly(A) tail consists of multiple adenosine monophosphates; in other words, it is a stretch of RNA that has only adenine bases. In eu ...
sites. A sequence identified as a putative gene must be confirmed by further evidence, such as similarity to cDNA or EST sequences from the same organism, similarity of the predicted protein sequence to known proteins, association with promoter sequences, or evidence that mutating the sequence produces an observable phenotype.
Rosetta stone approach
The Rosetta stone approach is a computational method for de-novo protein function prediction. It is based on the hypothesis that some proteins involved in a given physiological process may exist as two separate genes in one organism and as a single gene in another. Genomes are scanned for sequences that are independent in one organism and in a single open reading frame in another. If two genes have fused, it is predicted that they have similar biological functions that make such co-regulation advantageous.Bioinformatics methods for Functional genomics
Because of the large quantity of data produced by these techniques and the desire to find biologically meaningful patterns,bioinformatics
Bioinformatics () is an interdisciplinary field that develops methods and software tools for understanding biological data, in particular when the data sets are large and complex. As an interdisciplinary field of science, bioinformatics combin ...
is crucial to analysis of functional genomics data. Examples of techniques in this class are data clustering
Cluster analysis or clustering is the task of grouping a set of objects in such a way that objects in the same group (called a cluster) are more similar (in some sense) to each other than to those in other groups (clusters). It is a main task of ...
or principal component analysis
Principal component analysis (PCA) is a popular technique for analyzing large datasets containing a high number of dimensions/features per observation, increasing the interpretability of data while preserving the maximum amount of information, and ...
for unsupervised machine learning
Machine learning (ML) is a field of inquiry devoted to understanding and building methods that 'learn', that is, methods that leverage data to improve performance on some set of tasks. It is seen as a part of artificial intelligence.
Machine ...
(class detection) as well as artificial neural network
Artificial neural networks (ANNs), usually simply called neural networks (NNs) or neural nets, are computing systems inspired by the biological neural networks that constitute animal brains.
An ANN is based on a collection of connected units ...
s or support vector machine
In machine learning, support vector machines (SVMs, also support vector networks) are supervised learning models with associated learning algorithms that analyze data for classification and regression analysis. Developed at AT&T Bell Laboratories ...
s for supervised machine learning (class prediction, classification Classification is a process related to categorization, the process in which ideas and objects are recognized, differentiated and understood.
Classification is the grouping of related facts into classes.
It may also refer to:
Business, organizat ...
). Functional enrichment analysis is used to determine the extent of over- or under-expression (positive- or negative- regulators in case of RNAi screens) of functional categories relative to a background sets. Gene ontology
The Gene Ontology (GO) is a major bioinformatics initiative to unify the representation of gene and gene product attributes across all species. More specifically, the project aims to: 1) maintain and develop its controlled vocabulary of gene and g ...
based enrichment analysis are provided by DAVID
David (; , "beloved one") (traditional spelling), , ''Dāwūd''; grc-koi, Δαυΐδ, Dauíd; la, Davidus, David; gez , ዳዊት, ''Dawit''; xcl, Դաւիթ, ''Dawitʿ''; cu, Давíдъ, ''Davidŭ''; possibly meaning "beloved one". w ...
and gene set enrichment analysis
Gene set enrichment analysis (GSEA) (also called functional enrichment analysis or pathway enrichment analysis) is a method to identify classes of genes or proteins that are over-represented in a large set of genes or proteins, and may have an ass ...
(GSEA), pathway based analysis by Ingenuity and Pathway studio and protein complex based analysis by COMPLEAT.
 New computational methods have been developed for understanding the results of a deep mutational scanning experiment. 'phydms' compares the result of a deep mutational scanning experiment to a phylogenetic tree. This allows the user to infer if the selection process in nature applies similar constraints on a protein as the results of the deep mutational scan indicate. This may allow an experimenter to choose between different experimental conditions based on how well they reflect nature. Deep mutational scanning has also been used to infer protein-protein interactions. The authors used a thermodynamic model to predict the effects of mutations in different parts of a dimer. Deep mutational structure can also be used to infer protein structure. Strong positive epistasis between two mutations in a deep mutational scan can be indicative of two parts of the protein that are close to each other in 3-D space. This information can then be used to infer protein structure. A proof of principle of this approach was shown by two groups using the protein GB1.
Results from MPRA experiments have required machine learning approaches to interpret the data. A gapped k-mer SVM model has been used to infer the kmers that are enriched within cis-regulatory sequences with high activity compared to sequences with lower activity. These models provide high predictive power. Deep learning and random forest approaches have also been used to interpret the results of these high-dimensional experiments. These models are beginning to help develop a better understanding of non-coding DNA function towards gene-regulation.
New computational methods have been developed for understanding the results of a deep mutational scanning experiment. 'phydms' compares the result of a deep mutational scanning experiment to a phylogenetic tree. This allows the user to infer if the selection process in nature applies similar constraints on a protein as the results of the deep mutational scan indicate. This may allow an experimenter to choose between different experimental conditions based on how well they reflect nature. Deep mutational scanning has also been used to infer protein-protein interactions. The authors used a thermodynamic model to predict the effects of mutations in different parts of a dimer. Deep mutational structure can also be used to infer protein structure. Strong positive epistasis between two mutations in a deep mutational scan can be indicative of two parts of the protein that are close to each other in 3-D space. This information can then be used to infer protein structure. A proof of principle of this approach was shown by two groups using the protein GB1.
Results from MPRA experiments have required machine learning approaches to interpret the data. A gapped k-mer SVM model has been used to infer the kmers that are enriched within cis-regulatory sequences with high activity compared to sequences with lower activity. These models provide high predictive power. Deep learning and random forest approaches have also been used to interpret the results of these high-dimensional experiments. These models are beginning to help develop a better understanding of non-coding DNA function towards gene-regulation.
Consortium projects focused on Functional Genomics
The ENCODE project
TheENCODE
The Encyclopedia of DNA Elements (ENCODE) is a public research project which aims to identify functional elements in the human genome.
ENCODE also supports further biomedical research by "generating community resources of genomics data, software ...
(Encyclopedia of DNA elements) project is an in-depth analysis of the human genome whose goal is to identify all the functional elements of genomic DNA, in both coding and non-coding regions. Important results include evidence from genomic tiling arrays that most nucleotides are transcribed as coding transcripts, non-coding RNAs, or random transcripts, the discovery of additional transcriptional regulatory sites, further elucidation of chromatin-modifying mechanisms.
The Genotype-Tissue Expression (GTEx) project
 The GTEx project is a human genetics project aimed at understanding the role of genetic variation in shaping variation in the transcriptome across tissues. The project has collected a variety of tissue samples (> 50 different tissues) from more than 700 post-mortem donors. This has resulted in the collection of >11,000 samples. GTEx has helped understand the tissue-sharing and tissue-specificity of
The GTEx project is a human genetics project aimed at understanding the role of genetic variation in shaping variation in the transcriptome across tissues. The project has collected a variety of tissue samples (> 50 different tissues) from more than 700 post-mortem donors. This has resulted in the collection of >11,000 samples. GTEx has helped understand the tissue-sharing and tissue-specificity of eQTL
Expression quantitative trait loci (eQTLs) are genomic loci that explain variation in expression levels of mRNAs.
Distant and local, trans- and cis-eQTLs, respectively
An expression quantitative trait is an amount of an mRNA transcript or a pr ...
s. The genomic resource was developed to "enrich our understanding of how differences in our DNA sequence contribute to health and disease."
See also
*Systems biology
Systems biology is the computational modeling, computational and mathematical analysis and modeling of complex biological systems. It is a biology-based interdisciplinary field of study that focuses on complex interactions within biological syst ...
*Structural genomics
Structural genomics seeks to describe the 3-dimensional structure of every protein encoded by a given genome. This genome-based approach allows for a high-throughput method of structure determination by a combination of experimental and modeling ...
*Comparative genomics
Comparative genomics is a field of biological research in which the genomic features of different organisms are compared. The genomic features may include the DNA sequence, genes, gene order, regulatory sequences, and other genomic structural ...
*Pharmacogenomics
Pharmacogenomics is the study of the role of the genome in drug response. Its name ('' pharmaco-'' + ''genomics'') reflects its combining of pharmacology and genomics. Pharmacogenomics analyzes how the genetic makeup of an individual affects the ...
* MGED Society
*Epigenetics
In biology, epigenetics is the study of stable phenotypic changes (known as ''marks'') that do not involve alterations in the DNA sequence. The Greek prefix '' epi-'' ( "over, outside of, around") in ''epigenetics'' implies features that are " ...
*Bioinformatics
Bioinformatics () is an interdisciplinary field that develops methods and software tools for understanding biological data, in particular when the data sets are large and complex. As an interdisciplinary field of science, bioinformatics combin ...
* Epistasis and functional genomics
* Synthetic viability
*Protein function prediction
Protein function prediction methods are techniques that bioinformatics researchers use to assign biological or biochemical roles to proteins. These proteins are usually ones that are poorly studied or predicted based on genomic sequence data. Thes ...
References
External links
European Science Foundation Programme on Frontiers of Functional Genomics
— Integrated Functional Genomics in Mutant Mouse Models
Nature insights: functional genomics
{{DEFAULTSORT:Functional Genomics Molecular biology Genomics