Eukaryotic DNA Replication on:
[Wikipedia]
[Google]
[Amazon]
 Eukaryotic DNA replication is a conserved mechanism that restricts
Eukaryotic DNA replication is a conserved mechanism that restricts
 The Mcm proteins on chromatin form a head-to-head double hexamer with the two rings slightly tilted, twisted and off-centred to create a kink in the central channel where the bound DNA is captured at the interface of the two rings. Each hexameric Mcm2-7 ring first serves as the scaffold for the assembly of the replisome and then as the core of the catalytic CMG (Cdc45-MCM-GINS) helicase, which is a main component of the replisome. Each Mcm protein is highly related to all others, but unique sequences distinguishing each of the subunit types are conserved across eukaryotes. All eukaryotes have exactly six Mcm protein analogs that each fall into one of the existing classes (Mcm2-7), indicating that each Mcm protein has a unique and important function.
Minichromosome maintenance proteins are required for DNA helicase activity. Inactivation of any of the six Mcm proteins during S phase irreversibly prevents further progression of the replication fork suggesting that the helicase cannot be recycled and must be assembled at replication origins. Along with the minichromosome maintenance protein complex helicase activity, the complex also has associated ATPase activity. Studies have shown that within the Mcm protein complex are specific catalytic pairs of Mcm proteins that function together to coordinate ATP hydrolysis. These studies, confirmed by cryo-EM structures of the Mcm2-7 complexes, showed that the Mcm complex is a hexamer with subunits arranged in a ring in the order of Mcm2-Mcm6-Mcm4-Mcm7-Mcm3-Mcm5-. Both members of each catalytic pair contribute to the conformation that allows ATP binding and hydrolysis and the mixture of active and inactive subunits presumably allows the Mcm hexameric complex to complete ATP binding and hydrolysis as a whole to create a coordinated ATPase activity.
The
The Mcm proteins on chromatin form a head-to-head double hexamer with the two rings slightly tilted, twisted and off-centred to create a kink in the central channel where the bound DNA is captured at the interface of the two rings. Each hexameric Mcm2-7 ring first serves as the scaffold for the assembly of the replisome and then as the core of the catalytic CMG (Cdc45-MCM-GINS) helicase, which is a main component of the replisome. Each Mcm protein is highly related to all others, but unique sequences distinguishing each of the subunit types are conserved across eukaryotes. All eukaryotes have exactly six Mcm protein analogs that each fall into one of the existing classes (Mcm2-7), indicating that each Mcm protein has a unique and important function.
Minichromosome maintenance proteins are required for DNA helicase activity. Inactivation of any of the six Mcm proteins during S phase irreversibly prevents further progression of the replication fork suggesting that the helicase cannot be recycled and must be assembled at replication origins. Along with the minichromosome maintenance protein complex helicase activity, the complex also has associated ATPase activity. Studies have shown that within the Mcm protein complex are specific catalytic pairs of Mcm proteins that function together to coordinate ATP hydrolysis. These studies, confirmed by cryo-EM structures of the Mcm2-7 complexes, showed that the Mcm complex is a hexamer with subunits arranged in a ring in the order of Mcm2-Mcm6-Mcm4-Mcm7-Mcm3-Mcm5-. Both members of each catalytic pair contribute to the conformation that allows ATP binding and hydrolysis and the mixture of active and inactive subunits presumably allows the Mcm hexameric complex to complete ATP binding and hydrolysis as a whole to create a coordinated ATPase activity.
The
 The formation of the pre-replicative complex (pre-RC) marks the potential sites for the initiation of DNA replication. Consistent with the minichromosome maintenance complex encircling double stranded DNA, formation of the pre-RC does not lead to the immediate unwinding of origin DNA or the recruitment of DNA polymerases. Instead, the pre-RC that is formed during the G1 of the cell cycle is only activated to unwind the DNA and initiate replication after the cells pass from the G1 to the S phase of the cell cycle.
Once the initiation complex is formed and the cells pass into the S phase, the complex then becomes a replisome. The eukaryotic replisome complex is responsible for coordinating DNA replication. Replication on the leading and lagging strands is performed by DNA polymerase ε and DNA polymerase δ. Many replisome factors including Claspin, And1, replication factor C clamp loader and the fork protection complex are responsible for regulating polymerase functions and coordinating DNA synthesis with the unwinding of the template strand by Cdc45-Mcm-GINS complex. As the DNA is unwound the twist number decreases. To compensate for this the
The formation of the pre-replicative complex (pre-RC) marks the potential sites for the initiation of DNA replication. Consistent with the minichromosome maintenance complex encircling double stranded DNA, formation of the pre-RC does not lead to the immediate unwinding of origin DNA or the recruitment of DNA polymerases. Instead, the pre-RC that is formed during the G1 of the cell cycle is only activated to unwind the DNA and initiate replication after the cells pass from the G1 to the S phase of the cell cycle.
Once the initiation complex is formed and the cells pass into the S phase, the complex then becomes a replisome. The eukaryotic replisome complex is responsible for coordinating DNA replication. Replication on the leading and lagging strands is performed by DNA polymerase ε and DNA polymerase δ. Many replisome factors including Claspin, And1, replication factor C clamp loader and the fork protection complex are responsible for regulating polymerase functions and coordinating DNA synthesis with the unwinding of the template strand by Cdc45-Mcm-GINS complex. As the DNA is unwound the twist number decreases. To compensate for this the

 Termination of eukaryotic DNA replication requires different processes depending on whether the chromosomes are circular or linear. Unlike linear molecules, circular chromosomes are able to replicate the entire molecule. However, the two DNA molecules will remain linked together. This issue is handled by decatenation of the two DNA molecules by a
Termination of eukaryotic DNA replication requires different processes depending on whether the chromosomes are circular or linear. Unlike linear molecules, circular chromosomes are able to replicate the entire molecule. However, the two DNA molecules will remain linked together. This issue is handled by decatenation of the two DNA molecules by a
SciencedirectDOI
/ref>
 DNA replication is a tightly orchestrated process that is controlled within the context of the
DNA replication is a tightly orchestrated process that is controlled within the context of the
 Eukaryotic DNA must be tightly compacted in order to fit within the confined space of the nucleus. Chromosomes are packaged by wrapping 147 nucleotides around an octamer of
Eukaryotic DNA must be tightly compacted in order to fit within the confined space of the nucleus. Chromosomes are packaged by wrapping 147 nucleotides around an octamer of
 Eukaryotic DNA replication is a conserved mechanism that restricts
Eukaryotic DNA replication is a conserved mechanism that restricts DNA replication
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all living organisms acting as the most essential part for biological inheritanc ...
to once per cell cycle. Eukaryotic DNA replication of chromosomal
A chromosome is a long DNA molecule with part or all of the genetic material of an organism. In most chromosomes the very long thin DNA fibers are coated with packaging proteins; in eukaryotic cells the most important of these proteins are ...
DNA is central for the duplication of a cell
Cell most often refers to:
* Cell (biology), the functional basic unit of life
Cell may also refer to:
Locations
* Monastic cell, a small room, hut, or cave in which a religious recluse lives, alternatively the small precursor of a monastery w ...
and is necessary for the maintenance of the eukaryotic genome
In the fields of molecular biology and genetics, a genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding ge ...
.
DNA replication is the action of DNA polymerases
A DNA polymerase is a member of a family of enzymes that catalyze the synthesis of DNA molecules from nucleoside triphosphates, the molecular precursors of DNA. These enzymes are essential for DNA replication and usually work in groups to create ...
synthesizing a DNA strand complementary to the original template strand. To synthesize DNA, the double-stranded DNA is unwound by DNA helicases
Helicases are a class of enzymes thought to be vital to all organisms. Their main function is to unpack an organism's genetic material. Helicases are motor proteins that move directionally along a nucleic acid phosphodiester backbone, separating ...
ahead of polymerases, forming a replication fork containing two single-stranded templates. Replication processes permit the copying of a single DNA double helix into two DNA helices, which are divided into the daughter cells at mitosis
In cell biology, mitosis () is a part of the cell cycle in which replicated chromosomes are separated into two new nuclei. Cell division by mitosis gives rise to genetically identical cells in which the total number of chromosomes is mainta ...
. The major enzymatic functions carried out at the replication fork are well conserved from prokaryotes
A prokaryote () is a single-celled organism that lacks a nucleus and other membrane-bound organelles. The word ''prokaryote'' comes from the Greek πρό (, 'before') and κάρυον (, 'nut' or 'kernel').Campbell, N. "Biology:Concepts & Connec ...
to eukaryotes
Eukaryotes () are organisms whose cells have a nucleus. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the three domains of life. Bacte ...
, but the replication machinery in eukaryotic DNA replication is a much larger complex, coordinating many proteins at the site of replication, forming the replisome
The replisome is a complex molecular machine that carries out replication of DNA. The replisome first unwinds double stranded DNA into two single strands. For each of the resulting single strands, a new complementary sequence of DNA is synthe ...
.
The replisome is responsible for copying the entirety of genomic
Genomics is an interdisciplinary field of biology focusing on the structure, function, evolution, mapping, and editing of genomes. A genome is an organism's complete set of DNA, including all of its genes as well as its hierarchical, three-dim ...
DNA in each proliferative cell. This process allows for the high-fidelity passage of hereditary/genetic information from parental cell to daughter cell and is thus essential to all organisms. Much of the cell cycle
The cell cycle, or cell-division cycle, is the series of events that take place in a cell that cause it to divide into two daughter cells. These events include the duplication of its DNA (DNA replication) and some of its organelles, and subs ...
is built around ensuring that DNA replication occurs without errors.
In G1 phase of the cell cycle, many of the DNA replication regulatory processes are initiated. In eukaryotes, the vast majority of DNA synthesis
DNA synthesis is the natural or artificial creation of deoxyribonucleic acid (DNA) molecules. DNA is a macromolecule made up of nucleotide units, which are linked by covalent bonds and hydrogen bonds, in a repeating structure. DNA synthesis occur ...
occurs during S phase
S phase (Synthesis Phase) is the phase of the cell cycle in which DNA is replicated, occurring between G1 phase and G2 phase. Since accurate duplication of the genome is critical to successful cell division, the processes that occur during ...
of the cell cycle, and the entire genome must be unwound and duplicated to form two daughter copies. During G2, any damaged DNA or replication errors are corrected. Finally, one copy of the genomes is segregated to each daughter cell at mitosis or M phase. These daughter copies each contain one strand from the parental duplex DNA and one nascent antiparallel strand.
This mechanism is conserved from prokaryotes to eukaryotes and is known as semiconservative
Semiconservative replication describe the mechanism of DNA replication in all known cells. DNA replication occurs on multiple origins of replication along the DNA template strand (antinsense strand). As the DNA double helix is unwound by helicase, ...
DNA replication. The process of semiconservative replication for the site of DNA replication is a fork-like DNA structure, the replication fork, where the DNA helix is open, or unwound, exposing unpaired DNA nucleotides
Nucleotides are organic molecules consisting of a nucleoside and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of which are essential biomolecules w ...
for recognition and base pairing for the incorporation
of free nucleotides into double-stranded DNA.
Initiation
Initiation of eukaryotic DNA replication is the first stage of DNA synthesis where the DNA double helix is unwound and an initial priming event by DNA polymerase α occurs on the leading strand. The priming event on the lagging strand establishes a replication fork. Priming of the DNA helix consists of synthesis of an RNA primer to allow DNA synthesis by DNA polymerase α. Priming occurs once at the origin on the leading strand and at the start of each Okazaki fragment on the lagging strand. DNA replication is initiated from specific sequences calledorigins of replication
The origin of replication (also called the replication origin) is a particular sequence in a genome at which replication is initiated. Propagation of the genetic material between generations requires timely and accurate duplication of DNA by semi ...
, and eukaryotic cells have multiple replication origins. To initiate DNA replication, multiple replicative proteins assemble on and dissociate from these replicative origins. The individual factors described below work together to direct the formation of the pre-replication complex
A pre-replication complex (pre-RC) is a protein complex that forms at the origin of replication during the initiation step of DNA replication. Formation of the pre-RC is required for DNA replication to occur. Complete and faithful replication of ...
(pre-RC), a key intermediate in the replication initiation process.
Association of the origin recognition complex
In molecular biology, origin recognition complex (ORC) is a multi-subunit DNA binding complex (6 subunits) that binds in all eukaryotes and archaea in an ATP-dependent manner to origins of replication. The subunits of this complex are encoded ...
(ORC) with a replication origin recruits the cell division cycle 6 protein (Cdc6) to form a platform for the loading of the minichromosome maintenance (Mcm 2-7) complex proteins, facilitated by the chromatin licensing and DNA replication factor 1 protein (Cdt1). The ORC, Cdc6, and Cdt1 together are required for the stable association of the Mcm2-7 complex with replicative origins during G1 phase of the cell cycle.
Pre-replicative complex
Eukaryotic origins of replication control the formation of a number of protein complexes that lead to the assembly of two bidirectional DNA replication forks. These events are initiated by the formation of thepre-replication complex
A pre-replication complex (pre-RC) is a protein complex that forms at the origin of replication during the initiation step of DNA replication. Formation of the pre-RC is required for DNA replication to occur. Complete and faithful replication of ...
(pre-RC) at the origins of replication. This process takes place in the G1 stage of the cell cycle. The pre-RC formation involves the ordered assembly of many replication factors including the origin recognition complex (ORC), Cdc6 protein, Cdt1 protein, and minichromosome maintenance proteins (Mcm2-7). Once the pre-RC is formed, activation of the complex is triggered by two kinases
In biochemistry, a kinase () is an enzyme that catalysis, catalyzes the transfer of phosphate groups from High-energy phosphate, high-energy, phosphate-donating molecules to specific Substrate (biochemistry), substrates. This process is known as ...
, cyclin-dependent kinase 2
Cyclin-dependent kinase 2, also known as cell division protein kinase 2, or Cdk2, is an enzyme that in humans is encoded by the ''CDK2'' gene. The protein encoded by this gene is a member of the cyclin-dependent kinase family of Ser/Thr protein ...
(CDK) and Dbf4-dependent kinase (DDK) that help transition the pre-RC to the initiation complex prior to the initiation of DNA replication. This transition involves the ordered assembly of additional replication factors to unwind the DNA and accumulate the multiple eukaryotic DNA polymerases around the unwound DNA. Central to the question of how bidirectional replication forks are established at replication origins is the mechanism by which ORC recruits two head-to-head Mcm2-7 complexes to every replication origin to form the pre-replication complex.
Origin recognition complex
The first step in the assembly of the pre-replication complex (pre-RC) is the binding of theorigin recognition complex
In molecular biology, origin recognition complex (ORC) is a multi-subunit DNA binding complex (6 subunits) that binds in all eukaryotes and archaea in an ATP-dependent manner to origins of replication. The subunits of this complex are encoded ...
(ORC) to the replication origin. In late mitosis, Cdc6 protein joins the bound ORC followed by the binding of the Cdt1-Mcm2-7 complex. ORC, Cdc6, and Cdt1 are all required to load the six protein minichromosome maintenance (Mcm 2-7) complex onto the DNA. The ORC is a six-subunit, Orc1p-6, protein complex that selects the replicative origin sites on DNA for initiation of replication and ORC binding to chromatin is regulated through the cell cycle. Generally, the function and size of the ORC subunits are conserved throughout many eukaryotic genomes with the difference being their diverged DNA binding sites.
The most widely studied origin recognition complex is that of ''Saccharomyces cerevisiae
''Saccharomyces cerevisiae'' () (brewer's yeast or baker's yeast) is a species of yeast (single-celled fungus microorganisms). The species has been instrumental in winemaking, baking, and brewing since ancient times. It is believed to have been o ...
'' or yeast which is known to bind to the autonomously replicating sequence An autonomously replicating sequence (ARS) contains the origin of replication in the yeast genome. It contains four regions (A, B1, B2, and B3), named in order of their effect on plasmid stability. The A-Domain is highly conserved, any mutation abol ...
(ARS). The ''S. cerevisiae'' ORC interacts specifically with both the A and B1 elements of yeast origins of replication, spanning a region of 30 base pairs
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA ...
. The binding to these sequences requires ATP
ATP may refer to:
Companies and organizations
* Association of Tennis Professionals, men's professional tennis governing body
* American Technical Publishers, employee-owned publishing company
* ', a Danish pension
* Armenia Tree Project, non ...
.
The atomic structure of the ''S. cerevisiae'' ORC bound to ARS DNA has been determined. Orc1, Orc2, Orc3, Orc4, and Orc5 encircle the A element by means of two types of interactions, base non-specific and base-specific, that bend the DNA at the A element. All five subunits contact the sugar phosphate backbone at multiple points of the A element to form a tight grip without base specificity. Orc1 and Orc2 contact the minor groove of the A element while a winged helix domain of Orc4 contacts the methyl groups of the invariant Ts in the major groove of the A element via an insertion helix (IH). The absence of this IH in metazoans explains the lack of sequence specificity in human ORC. Removing the IH from the ''Sc''ORC causes it to lose its specificity for the A element, and to bind promiscuously and preferentially (83%) to promoter regions. The ARS DNA is also bent at the B1 element through interactions with Orc2, Orc5 and Orc6. The bending of origin DNA by ORC appears to be evolutionarily conserved suggesting that it may be required for the Mcm2-7 complex loading mechanism.
When the ORC binds to DNA at replication origins, it serves as a scaffold for the assembly of other key initiation factors of the pre-replicative complex. This pre-replicative complex assembly during the G1 stage of the cell cycle is required prior to the activation of DNA replication during the S phase. The removal of at least part of the complex (Orc1) from the chromosome at metaphase
Metaphase ( and ) is a stage of mitosis in the eukaryotic cell cycle in which chromosomes are at their second-most condensed and coiled stage (they are at their most condensed in anaphase). These chromosomes, carrying genetic information, align ...
is part of the regulation of mammalian ORC to ensure that the pre-replicative complex formation prior to the completion of metaphase is eliminated.
Cdc6 protein
Binding of the cell division cycle 6 (Cdc6) protein to the origin recognition complex (ORC) is an essential step in the assembly of the pre-replication complex (pre-RC) at the origins of replication. Cdc6 binds to the ORC on DNA in an ATP-dependent manner, which induces a change in the pattern of origin binding that requires Orc1ATPase
ATPases (, Adenosine 5'-TriPhosphatase, adenylpyrophosphatase, ATP monophosphatase, triphosphatase, SV40 T-antigen, ATP hydrolase, complex V (mitochondrial electron transport), (Ca2+ + Mg2+)-ATPase, HCO3−-ATPase, adenosine triphosphatase) are ...
. Cdc6 requires ORC in order to associate with chromatin and is in turn required for the Cdt1-Mcm2-7 heptamer to bind to the chromatin. The ORC-Cdc6 complex forms a ring-shaped structure and is analogous to other ATP-dependent protein machines. The levels and activity of Cdc6 regulate the frequency with which the origins of replication are utilized during the cell cycle.
Cdt1 protein
The chromatin licensing and DNA replication factor 1 (Cdt1) protein is required for the licensing of chromatin for DNA replication. In ''S. cerevisiae'', Cdt1 facilitates the loading of the Mcm2-7 complex one at a time onto the chromosome by stabilising the left-handed open-ring structure of the Mcm2-7 single hexamer. Cdt1 has been shown to associate with theC terminus
The C-terminus (also known as the carboxyl-terminus, carboxy-terminus, C-terminal tail, C-terminal end, or COOH-terminus) is the end of an amino acid chain (protein or polypeptide), terminated by a free carboxyl group (-COOH). When the protein is ...
of Cdc6 to cooperatively promote the association of Mcm proteins to the chromatin. The cryo-EM structure of the OCCM (ORC-Cdc6-Cdt1-MCM) complex shows that the Cdt1-CTD interacts with the Mcm6-WHD. In metazoans, Cdt1 activity during the cell cycle is tightly regulated by its association with the protein geminin
Geminin, DNA replication inhibitor, also known as GMNN, is a protein in humans encoded by the ''GMNN'' gene. A nuclear protein present in most eukaryotes and highly conserved across species, numerous functions have been elucidated for geminin inc ...
, which both inhibits Cdt1 activity during S phase in order to prevent re-replication of DNA and prevents it from ubiquitination
Ubiquitin is a small (8.6 kDa) regulatory protein found in most tissues of eukaryotic organisms, i.e., it is found ''ubiquitously''. It was discovered in 1975 by Gideon Goldstein and further characterized throughout the late 1970s and 1980s. Fo ...
and subsequent proteolysis
Proteolysis is the breakdown of proteins into smaller polypeptides or amino acids. Uncatalysed, the hydrolysis of peptide bonds is extremely slow, taking hundreds of years. Proteolysis is typically catalysed by cellular enzymes called protease ...
.
Minichromosome maintenance protein complex
The minichromosome maintenance (Mcm) proteins were named after a genetic screen for DNA replication initiation mutants in ''S. cerevisiae'' that affect plasmid stability in an ARS-specific manner''.'' Mcm2, Mcm3, Mcm4, Mcm5, Mcm6 and Mcm7 form a hexameric complex that has an open-ring structure with a gap between Mcm2 and Mcm5. The assembly of the Mcm proteins onto chromatin requires the coordinated function of the origin recognition complex (ORC), Cdc6, and Cdt1. Once the Mcm proteins have been loaded onto the chromatin, ORC and Cdc6 can be removed from the chromatin without preventing subsequent DNA replication. This observation suggests that the primary role of the pre-replication complex is to correctly load the Mcm proteins. The Mcm proteins on chromatin form a head-to-head double hexamer with the two rings slightly tilted, twisted and off-centred to create a kink in the central channel where the bound DNA is captured at the interface of the two rings. Each hexameric Mcm2-7 ring first serves as the scaffold for the assembly of the replisome and then as the core of the catalytic CMG (Cdc45-MCM-GINS) helicase, which is a main component of the replisome. Each Mcm protein is highly related to all others, but unique sequences distinguishing each of the subunit types are conserved across eukaryotes. All eukaryotes have exactly six Mcm protein analogs that each fall into one of the existing classes (Mcm2-7), indicating that each Mcm protein has a unique and important function.
Minichromosome maintenance proteins are required for DNA helicase activity. Inactivation of any of the six Mcm proteins during S phase irreversibly prevents further progression of the replication fork suggesting that the helicase cannot be recycled and must be assembled at replication origins. Along with the minichromosome maintenance protein complex helicase activity, the complex also has associated ATPase activity. Studies have shown that within the Mcm protein complex are specific catalytic pairs of Mcm proteins that function together to coordinate ATP hydrolysis. These studies, confirmed by cryo-EM structures of the Mcm2-7 complexes, showed that the Mcm complex is a hexamer with subunits arranged in a ring in the order of Mcm2-Mcm6-Mcm4-Mcm7-Mcm3-Mcm5-. Both members of each catalytic pair contribute to the conformation that allows ATP binding and hydrolysis and the mixture of active and inactive subunits presumably allows the Mcm hexameric complex to complete ATP binding and hydrolysis as a whole to create a coordinated ATPase activity.
The
The Mcm proteins on chromatin form a head-to-head double hexamer with the two rings slightly tilted, twisted and off-centred to create a kink in the central channel where the bound DNA is captured at the interface of the two rings. Each hexameric Mcm2-7 ring first serves as the scaffold for the assembly of the replisome and then as the core of the catalytic CMG (Cdc45-MCM-GINS) helicase, which is a main component of the replisome. Each Mcm protein is highly related to all others, but unique sequences distinguishing each of the subunit types are conserved across eukaryotes. All eukaryotes have exactly six Mcm protein analogs that each fall into one of the existing classes (Mcm2-7), indicating that each Mcm protein has a unique and important function.
Minichromosome maintenance proteins are required for DNA helicase activity. Inactivation of any of the six Mcm proteins during S phase irreversibly prevents further progression of the replication fork suggesting that the helicase cannot be recycled and must be assembled at replication origins. Along with the minichromosome maintenance protein complex helicase activity, the complex also has associated ATPase activity. Studies have shown that within the Mcm protein complex are specific catalytic pairs of Mcm proteins that function together to coordinate ATP hydrolysis. These studies, confirmed by cryo-EM structures of the Mcm2-7 complexes, showed that the Mcm complex is a hexamer with subunits arranged in a ring in the order of Mcm2-Mcm6-Mcm4-Mcm7-Mcm3-Mcm5-. Both members of each catalytic pair contribute to the conformation that allows ATP binding and hydrolysis and the mixture of active and inactive subunits presumably allows the Mcm hexameric complex to complete ATP binding and hydrolysis as a whole to create a coordinated ATPase activity.
The nuclear localization A nuclear localization signal ''or'' sequence (NLS) is an amino acid sequence that 'tags' a protein for import into the cell nucleus by nuclear transport. Typically, this signal consists of one or more short sequences of positively charged lysines o ...
of the minichromosome maintenance proteins is regulated in budding yeast cells. The Mcm proteins are present in the nucleus
Nucleus ( : nuclei) is a Latin word for the seed inside a fruit. It most often refers to:
*Atomic nucleus, the very dense central region of an atom
*Cell nucleus, a central organelle of a eukaryotic cell, containing most of the cell's DNA
Nucle ...
in G1 stage and S phase of the cell cycle, but are exported to the cytoplasm
In cell biology, the cytoplasm is all of the material within a eukaryotic cell, enclosed by the cell membrane, except for the cell nucleus. The material inside the nucleus and contained within the nuclear membrane is termed the nucleoplasm. The ...
during the G2 stage and M phase. A complete and intact six subunit Mcm complex is required to enter into the cell nucleus. In ''S. cerevisiae'', nuclear export
A nuclear export signal (NES) is a short target peptide containing 4 hydrophobic residues in a protein that targets it for export from the cell nucleus to the cytoplasm through the nuclear pore complex using nuclear transport. It has the opposite ...
is promoted by cyclin-dependent kinase (CDK) activity. Mcm proteins that are associated with chromatin are protected from CDK export machinery due to the lack of accessibility to CDK.
Initiation complex
During the G1 stage of the cell cycle, the replication initiation factors, origin recognition complex (ORC), Cdc6, Cdt1, and minichromosome maintenance (Mcm) protein complex, bind sequentially to DNA to form the pre-replication complex (pre-RC). At the transition of the G1 stage to the S phase of the cell cycle, S phase–specific cyclin-dependent protein kinase (CDK) and Cdc7/Dbf4 kinase (DDK) transform the pre-RC into an active replication fork. During this transformation, the pre-RC is disassembled with the loss of Cdc6, creating the initiation complex. In addition to the binding of the Mcm proteins, cell division cycle 45 (Cdc45) protein is also essential for initiating DNA replication. Studies have shown that Mcm is critical for the loading of Cdc45 onto chromatin and this complex containing both Mcm and Cdc45 is formed at the onset of the S phase of the cell cycle. Cdc45 targets the Mcm protein complex, which has been loaded onto the chromatin, as a component of the pre-RC at the origin of replication during the G1 stage of the cell cycle.Cdc45 protein
Cell division cycle 45 (Cdc45) protein is a critical component for the conversion of the pre-replicative complex to the initiation complex. The Cdc45 protein assembles at replication origins before initiation and is required for replication to begin in ''Saccharomyces cerevisiae'', and has an essential role during elongation. Thus, Cdc45 has central roles in both initiation and elongation phases of chromosomal DNA replication. Cdc45 associates with chromatin after the beginning of initiation in late G1 stage and during the S phase of the cell cycle. Cdc45 physically associates with Mcm5 and displays genetic interactions with five of the six members of the Mcm gene family and theORC2
Origin recognition complex subunit 2 is a protein that is encoded by the ORC2 (ORC2L) gene in humans.
Function
The origin recognition complex (ORC) is a highly conserved six subunits protein complex essential for the initiation of the DNA repl ...
gene. The loading of Cdc45 onto chromatin is critical for loading other various replication proteins, including DNA polymerase α, DNA polymerase ε, replication protein A
Replication protein A (RPA) is the major protein that binds to single-stranded DNA (ssDNA) in eukaryotic cells. In vitro, RPA shows a much higher affinity for ssDNA than RNA or double-stranded DNA. RPA is required in replication, recombinatio ...
(RPA) and proliferating cell nuclear antigen
Proliferating cell nuclear antigen (PCNA) is a DNA clamp that acts as a processivity factor for DNA polymerase δ in eukaryotic cells and is essential for replication. PCNA is a homotrimer and achieves its processivity by encircling the DNA, whe ...
(PCNA) onto chromatin.
Within a ''Xenopus'' nucleus-free system, it has been demonstrated that Cdc45 is required for the unwinding of plasmid DNA. The ''Xenopus'' nucleus-free system also demonstrates that DNA unwinding and tight RPA binding to chromatin occurs only in the presence of Cdc45.
Binding of Cdc45 to chromatin depends on Clb-Cdc28 kinase activity as well as functional Cdc6 and Mcm2, which suggests that Cdc45 associates with the pre-RC after activation of S-phase cyclin-dependent kinases (CDKs). As indicated by the timing and the CDK dependence, binding of Cdc45 to chromatin is crucial for commitment to initiation of DNA replication. During S phase, Cdc45 physically interacts with Mcm proteins on chromatin; however, dissociation of Cdc45 from chromatin is slower than that of the Mcm, which indicates that the proteins are released by different mechanisms.
GINS
The six minichromosome maintenance proteins and Cdc45 are essential during initiation and elongation for the movement of replication forks and for unwinding of the DNA. GINS are essential for the interaction of Mcm and Cdc45 at the origins of replication during initiation and then at DNA replication forks as the replisome progresses. The GINS complex is composed of four small proteins Sld5 (Cdc105), Psf1 (Cdc101), Psf2 (Cdc102) and Psf3 (Cdc103), GINS represents 'go, ichi, ni, san' which means '5, 1, 2, 3' in Japanese. Cdc45, Mcm2-7 and GINS together form the CMG helicase, the replicative helicase of the replisome. Although the Mcm2-7 complex alone has weak helicase activity Cdc45 and GINS are required for robust helicase activityMcm10
Mcm10
Protein MCM10 homolog is a protein that in humans is encoded by the ''MCM10'' gene. It is essential for activation of the Cdc45: Mcm2-7:GINS helicase, and thus required for proper DNA replication.
Function
The protein encoded by this gene is o ...
is essential for chromosome replication and interacts with the minichromosome maintenance 2-7 helicase that is loaded in an inactive form at origins of DNA replication. Mcm10 also chaperones the catalytic DNA polymerase α and helps stabilize the polymerase at replication forks.
DDK and CDK kinases
At the onset of S phase, the pre-replicative complex must be activated by two S phase-specific kinases in order to form an initiation complex at an origin of replication. One kinase is the Cdc7-Dbf4 kinase called Dbf4-dependent kinase (DDK) and the other iscyclin-dependent kinase
Cyclin-dependent kinases (CDKs) are the families of protein kinases first discovered for their role in regulating the cell cycle. They are also involved in regulating transcription, mRNA processing, and the differentiation of nerve cells. They a ...
(CDK). Chromatin-binding assays of Cdc45 in yeast and ''Xenopus'' have shown that a downstream event of CDK action is loading of Cdc45 onto chromatin. Cdc6
Cell division control protein 6 homolog is a protein that in humans is encoded by the ''CDC6'' gene.
The protein encoded by this gene is highly similar to Saccharomyces cerevisiae Cdc6, a protein essential for the initiation of DNA replication. ...
has been speculated to be a target of CDK action, because of the association between Cdc6 and CDK, and the CDK-dependent phosphorylation
In chemistry, phosphorylation is the attachment of a phosphate group to a molecule or an ion. This process and its inverse, dephosphorylation, are common in biology and could be driven by natural selection. Text was copied from this source, wh ...
of Cdc6. The CDK-dependent phosphorylation of Cdc6 has been considered to be required for entry into the S phase.
Both the catalytic subunits of DDK, Cdc7, and the activator protein, Dbf4, are conserved in eukaryotes and are required for the onset of S phase of the cell cycle. Both Dbf4 and Cdc7 are required for the loading of Cdc45 onto chromatin origins of replication. The target for binding of the DDK kinase is the chromatin-bound form of the Mcm complex. High resolution cryoEM structures showed that the Dbf4 subunit of DDK straddles across the hexamer interface of the DNA-bound MCM-DH, contacting Mcm2 of one hexamer and Mcm4/6 of the opposite hexamer. Mcm2, Mcm4 and Mcm6 are all substrates of phosphorylation by DDK but only the N-terminal serine/threonine-rich domain (NSD) of Mcm4 is an essential DDK target. Phosphorylation of the NSD leads to the activation of Mcm helicase activity.
Dpb11, Sld3, and Sld2 proteins
Sld3, Sld2, and Dpb11 interact with many replication proteins. Sld3 and Cdc45 form a complex that associated with the pre-RC at the early origins of replication even in the G11 phase and with the later origins of replication in the S phase in a mutually Mcm-dependent manner. Dpb11 and Sld2 interact with Polymerase ɛ and cross-linking experiments have indicated that Dpb11 and Polymerase ɛ coprecipitate in the S phase and associate with replication origins. Sld3 and Sld2 are phosphorylated by CDK, which enables the two replicative proteins to bind to Dpb11. Dpb11 had two pairs of BRCA1 C Terminus (BRCT) domains which are known as a phosphopeptide-binding domains. The N-terminal pair of the BRCT domains binds to phosphorylated Sld3, and the C-terminal pair binds to phosphorylated Sld2. Both of these interactions are essential for CDK-dependent activation of DNA budding in yeast. Dpb11 also interacts with GINS and participates in the initiation and elongation steps of chromosomal DNA replication. GINS are one of the replication proteins found at the replication forks and forms a complex with Cdc45 and Mcm. These phosphorylation-dependent interactions between Dpb11, Sld2, and Sld3 are essential for CDK-dependent activation of DNA replication, and by using cross-linking reagents within some experiments, a fragile complex was identified called the pre-loading complex (pre-LC). This complex contains Pol ɛ, GINS, Sld2, and Dpb11. The pre-LC is found to form before any association with the origins in a CDK-dependent and DDK-dependent manner and CDK activity regulates the initiation of DNA replication through the formation of the pre-LC.Elongation
writhe
In knot theory, there are several competing notions of the quantity writhe, or \operatorname. In one sense, it is purely a property of an oriented link diagram and assumes integer values. In another sense, it is a quantity that describes the amou ...
number increases, introducing positive supercoils in the DNA. These supercoils would cause DNA replication to halt if they were not removed. Topoisomerases
DNA topoisomerases (or topoisomerases) are enzymes that catalyze changes in the topological state of DNA, interconverting relaxed and supercoiled forms, linked (catenated) and unlinked species, and knotted and unknotted DNA. Topological issues i ...
are responsible for removing these supercoils ahead of the replication fork.
The replisome is responsible for copying the entire genomic DNA in each proliferative cell. The base pairing and chain formation reactions, which form the daughter helix, are catalyzed by DNA polymerases. These enzymes move along single-stranded DNA and allow for the extension of the nascent DNA strand by "reading" the template strand and allowing for incorporation of the proper purine
Purine is a heterocyclic compound, heterocyclic aromatic organic compound that consists of two rings (pyrimidine and imidazole) fused together. It is water-soluble. Purine also gives its name to the wider class of molecules, purines, which includ ...
nucleobases
Nucleobases, also known as ''nitrogenous bases'' or often simply ''bases'', are nitrogen-containing biological compounds that form nucleosides, which, in turn, are components of nucleotides, with all of these monomers constituting the basic b ...
, adenine
Adenine () ( symbol A or Ade) is a nucleobase (a purine derivative). It is one of the four nucleobases in the nucleic acid of DNA that are represented by the letters G–C–A–T. The three others are guanine, cytosine and thymine. Its derivati ...
and guanine
Guanine () ( symbol G or Gua) is one of the four main nucleobases found in the nucleic acids DNA and RNA, the others being adenine, cytosine, and thymine (uracil in RNA). In DNA, guanine is paired with cytosine. The guanine nucleoside is called ...
, and pyrimidine
Pyrimidine (; ) is an aromatic, heterocyclic, organic compound similar to pyridine (). One of the three diazines (six-membered heterocyclics with two nitrogen atoms in the ring), it has nitrogen atoms at positions 1 and 3 in the ring. The other ...
nucleobases, thymine
Thymine () ( symbol T or Thy) is one of the four nucleobases in the nucleic acid of DNA that are represented by the letters G–C–A–T. The others are adenine, guanine, and cytosine. Thymine is also known as 5-methyluracil, a pyrimidine nu ...
and cytosine
Cytosine () ( symbol C or Cyt) is one of the four nucleobases found in DNA and RNA, along with adenine, guanine, and thymine (uracil in RNA). It is a pyrimidine derivative, with a heterocyclic aromatic ring and two substituents attached (an am ...
. Activated free deoxyribonucleotides
A deoxyribonucleotide is a nucleotide that contains deoxyribose. They are the monomeric units of the informational biopolymer, deoxyribonucleic acid ( DNA). Each deoxyribonucleotide comprises three parts: a deoxyribose sugar ( monosaccharide), a n ...
exist in the cell as deoxyribonucleotide triphosphates (dNTPs). These free nucleotides are added to an exposed 3'-hydroxyl group on the last incorporated nucleotide. In this reaction, a pyrophosphate is released from the free dNTP, generating energy for the polymerization reaction and exposing the 5' monophosphate, which is then covalently bonded to the 3' oxygen. Additionally, incorrectly inserted nucleotides can be removed and replaced by the correct nucleotides in an energetically favorable reaction. This property is vital to proper proofreading and repair of errors that occur during DNA replication.
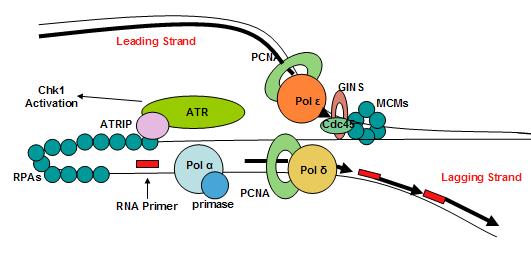
Replication fork
The replication fork is the junction between the newly separated template strands, known as the leading and lagging strands, and the double stranded DNA. Since duplex DNA is antiparallel, DNA replication occurs in opposite directions between the two new strands at the replication fork, but all DNA polymerases synthesize DNA in the 5' to 3' direction with respect to the newly synthesized strand. Further coordination is required during DNA replication. Two replicative polymerases synthesize DNA in opposite orientations. Polymerase ε synthesizes DNA on the "leading" DNA strand continuously as it is pointing in the same direction as DNA unwinding by the replisome. In contrast, polymerase δ synthesizes DNA on the "lagging" strand, which is the opposite DNA template strand, in a fragmented or discontinuous manner. The discontinuous stretches of DNA replication products on the lagging strand are known as Okazaki fragments and are about 100 to 200 bases in length at eukaryotic replication forks. The lagging strand usually contains longer stretches of single-stranded DNA that is coated with single-stranded binding proteins, which help stabilize the single-stranded templates by preventing a secondary structure formation. In eukaryotes, these single-stranded binding proteins are a heterotrimeric complex known asreplication protein A
Replication protein A (RPA) is the major protein that binds to single-stranded DNA (ssDNA) in eukaryotic cells. In vitro, RPA shows a much higher affinity for ssDNA than RNA or double-stranded DNA. RPA is required in replication, recombinatio ...
(RPA).
Each Okazaki fragment is preceded by an RNA primer, which is displaced by the procession of the next Okazaki fragment during synthesis. RNase H
Ribonuclease H (abbreviated RNase H or RNH) is a family of non-sequence-specific endonuclease enzymes that catalyze the cleavage of RNA in an RNA/ DNA substrate via a hydrolytic mechanism. Members of the RNase H family can be found in nearly a ...
recognizes the DNA:RNA hybrids that are created by the use of RNA primers and is responsible for removing these from the replicated strand, leaving behind a primer:template junction. DNA polymerase α, recognizes these sites and elongates the breaks left by primer removal. In eukaryotic cells, a small amount of the DNA segment immediately upstream of the RNA primer is also displaced, creating a flap structure. This flap is then cleaved by endonucleases. At the replication fork, the gap in DNA after removal of the flap is sealed by DNA ligase I
DNA ligase 1 is an enzyme that in humans is encoded by the ''LIG1'' gene. DNA ligase I is the only known eukaryotic DNA ligase involved in both DNA replication and repair, making it the most studied of the ligases.
Discovery
It was known that ...
, which repairs the nicks that are left between the 3'-OH and 5'phosphate of the newly synthesized strand. Owing to the relatively short nature of the eukaryotic Okazaki fragment, DNA replication synthesis occurring discontinuously on the lagging strand is less efficient and more time-consuming than leading-strand synthesis. DNA synthesis is complete once all RNA primers are removed and nicks are repaired.
Leading strand
During DNA replication, the replisome will unwind the parental duplex DNA into a two single-stranded DNA template replication fork in a 5' to 3' direction. The leading strand is the template strand that is being replicated in the same direction as the movement of the replication fork. This allows the newly synthesized strand complementary to the original strand to be synthesized 5' to 3' in the same direction as the movement of the replication fork. Once an RNA primer has been added by a primase to the 3' end of the leading strand, DNA synthesis will continue in a 3' to 5' direction with respect to the leading strand uninterrupted. DNA Polymerase ε will continuously add nucleotides to the template strand therefore making leading strand synthesis require only one primer and has uninterrupted DNA polymerase activity.Lagging strand
DNA replication on thelagging strand
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all living organisms acting as the most essential part for biological inheritance ...
is discontinuous. In lagging strand synthesis, the movement of DNA polymerase
A DNA polymerase is a member of a family of enzymes that catalyze the synthesis of DNA molecules from nucleoside triphosphates, the molecular precursors of DNA. These enzymes are essential for DNA replication and usually work in groups to create ...
in the opposite direction of the replication fork requires the use of multiple RNA primers. DNA polymerase will synthesize short fragments of DNA called Okazaki fragments
Okazaki fragments are short sequences of DNA nucleotides (approximately 150 to 200 base pairs long in eukaryotes) which are synthesized discontinuously and later linked together by the enzyme DNA ligase to create the lagging strand during DNA r ...
which are added to the 3' end of the primer. These fragments can be anywhere between 100–400 nucleotides long in eukaryotes.Watson J, Baker T, Bell S, Gann A, Levine M, Losick R, ''Molecular Biology of the Gene 6th Edition'', Pearson Education Inc, 2008.
At the end of Okazaki fragment synthesis, DNA polymerase δ runs into the previous Okazaki fragment and displaces its 5' end containing the RNA primer and a small segment of DNA. This generates an RNA-DNA single strand flap, which must be cleaved, and the nick between the two Okazaki fragments must be sealed by DNA ligase I. This process is known as Okazaki fragment maturation and can be handled in two ways: one mechanism processes short flaps, while the other deals with long flaps. DNA polymerase δ is able to displace up to 2 to 3 nucleotides of DNA or RNA ahead of its polymerization, generating a short "flap" substrate for Fen1
Flap endonuclease 1 is an enzyme that in humans is encoded by the ''FEN1'' gene.
Function
The protein encoded by this gene removes 5' overhanging "flaps" (or short sections of single stranded DNA that "hang off" because their nucleotide bases a ...
, which can remove nucleotides from the flap, one nucleotide at a time.
By repeating cycles of this process, DNA polymerase δ and Fen1 can coordinate the removal of RNA primers and leave a DNA nick at the lagging strand. It has been proposed that this iterative process is preferable to the cell because it is tightly
regulated and does not generate large flaps that need to be excised. In the event of deregulated Fen1/DNA polymerase δ activity, the cell uses an alternative mechanism to generate and process long flaps by using Dna2, which has both helicase and nuclease activities. The nuclease activity of Dna2 is required for removing these long flaps, leaving a shorter flap to be processed by Fen1. Electron microscopy
An electron microscope is a microscope that uses a beam of accelerated electrons as a source of illumination. As the wavelength of an electron can be up to 100,000 times shorter than that of visible light photons, electron microscopes have a hi ...
studies indicate that nucleosome loading on the lagging strand occurs very close to the site of synthesis. Thus, Okazaki fragment maturation is an efficient process that occurs immediately after the nascent DNA is synthesized.
Replicative DNA polymerases
After the replicative helicase has unwound the parental DNA duplex, exposing two single-stranded DNA templates, replicative polymerases are needed to generate two copies of the parental genome. DNA polymerase function is highly specialized and accomplish replication on specific templates and in narrow localizations. At the eukaryotic replication fork, there are three distinct replicative polymerase complexes that contribute to DNA replication: Polymerase α, Polymerase δ, and Polymerase ε. These three polymerases are essential for viability of the cell. Because DNA polymerases require a primer on which to begin DNA synthesis, polymerase α (Pol α) acts as a replicative primase. Pol α is associated with an RNA primase and this complex accomplishes the priming task by synthesizing a primer that contains a short 10 nucleotide stretch of RNA followed by 10 to 20 DNA bases. Importantly, this priming action occurs at replication initiation at origins to begin leading-strand synthesis and also at the 5' end of each Okazaki fragment on the lagging strand. However, Pol α is not able to continue DNA replication and must be replaced with another polymerase to continue DNA synthesis. Polymerase switching requires clamp loaders and it has been proven that normal DNA replication requires the coordinated actions of all three DNA polymerases: Pol α for priming synthesis, Pol ε for leading-strand replication, and the Pol δ, which is constantly loaded, for generating Okazaki fragments during lagging-strand synthesis. * Polymerase α (Pol α): Forms a complex with a small catalytic subunit (PriS) and a large noncatalytic (PriL) subunit. First, synthesis of an RNA primer allows DNA synthesis by DNA polymerase alpha. Occurs once at the origin on the leading strand and at the start of each Okazaki fragment on the lagging strand. Pri subunits act as a primase, synthesizing an RNA primer. DNA Pol α elongates the newly formed primer with DNA nucleotides. After around 20 nucleotides, elongation is taken over by Pol ε on the leading strand and Pol δ on the lagging strand. * Polymerase δ (Pol δ): Highly processive and has proofreading, 3'->5' exonuclease activity. ''In vivo,'' it is the main polymerase involved in both lagging strand ''and'' leading strand synthesis. * Polymerase ε (Pol ε): Highly processive and has proofreading, 3'->5' exonuclease activity. Highly related to pol δ, ''in vivo'' it functions mainly in error checking of pol δ.Cdc45–Mcm–GINS helicase complex
The DNAhelicases
Helicases are a class of enzymes thought to be vital to all organisms. Their main function is to unpack an organism's genetic material. Helicases are motor proteins that move directionally along a nucleic acid phosphodiester backbone, separating ...
and polymerases must remain in close contact at the replication fork. If unwinding occurs too far in advance of synthesis, large tracts of single-stranded DNA are exposed. This can activate DNA damage signaling or induce DNA repair processes. To thwart these problems, the eukaryotic replisome contains specialized proteins that are designed to regulate the helicase activity ahead of the replication fork. These proteins also provide docking sites for physical interaction between helicases and polymerases, thereby ensuring that duplex unwinding is coupled with DNA synthesis.
For DNA polymerases to function, the double-stranded DNA helix has to be unwound to expose two single-stranded DNA templates for replication. DNA helicases are responsible for unwinding the double-stranded DNA during chromosome replication. Helicases in eukaryotic cells are remarkably complex. The catalytic core of the helicase is composed of six minichromosome maintenance (Mcm2-7) proteins, forming a hexameric
In chemistry and biochemistry, an oligomer () is a molecule that consists of a few repeating units which could be derived, actually or conceptually, from smaller molecules, monomers.Quote: ''Oligomer molecule: A molecule of intermediate relative ...
ring. Away from DNA, the Mcm2-7 proteins form a single heterohexamer and are loaded in an inactive form at origins of DNA replication as a head-to-head double hexamers around double-stranded DNA. The Mcm proteins are recruited to replication origins then redistributed throughout the genomic DNA during S phase, indicative of their localization to the replication fork.
Loading of Mcm proteins can only occur during the G1 of the cell cycle, and the loaded complex is then activated during S phase by recruitment of the Cdc45 protein and the GINS complex to form the active Cdc45–Mcm–GINS (CMG) helicase at DNA replication forks. Mcm activity is required throughout the S phase for DNA replication. A variety of regulatory factors assemble around the CMG helicase to produce the ‘Replisome Progression Complex’ which associates with DNA polymerases to form the eukaryotic replisome, the structure of which is still quite poorly defined in comparison with its bacterial counterpart.
The isolated CMG helicase and Replisome Progression Complex contain a single Mcm protein ring complex suggesting that the loaded double hexamer of the Mcm proteins at origins might be broken into two single hexameric rings as part of the initiation process, with each Mcm protein complex ring forming the core of a CMG helicase at the two replication forks established from each origin. The full CMG complex is required for DNA unwinding, and the complex of CDC45-Mcm-GINS is the functional DNA helicase in eukaryotic cells.
Ctf4 and And1 proteins
The CMG complex interacts with the replisome through the interaction with Ctf4 and And1 proteins. Ctf4/And1 proteins interact with both the CMG complex and DNA polymerase α. Ctf4 is a polymerase α accessory factor, which is required for the recruitment of polymerase α to replication origins.Mrc1 and Claspin proteins
Mrc1/Claspin proteins couple leading-strand synthesis with the CMG complex helicase activity. Mrc1 interacts with polymerase ε as well as Mcm proteins. The importance of this direct link between the helicase and the leading-strand polymerase is underscored by results in cultured human cells, where Mrc1/Claspin is required for efficient replication fork progression. These results suggest that efficient DNA replication also requires the coupling of helicases and leading-strand synthesis...Proliferating cell nuclear antigen
DNA polymerases require additional factors to support DNA replication. DNA polymerases have a semiclosed 'hand' structure, which allows the polymerase to load onto the DNA and begin translocating. This structure permits DNA polymerase to hold the single-stranded DNA template, incorporate dNTPs at the active site, and release the newly formed double-stranded DNA. However, the structure of DNA polymerases does not allow a continuous stable interaction with the template DNA. To strengthen the interaction between the polymerase and the template DNA, DNA sliding clamps associate with the polymerase to promote theprocessivity
In molecular biology and biochemistry, processivity is an enzyme's ability to catalyze "consecutive reactions without releasing its substrate".
For example, processivity is the average number of nucleotides added by a polymerase enzyme, such as ...
of the replicative polymerase. In eukaryotes, the sliding clamp is a homotrimer ring structure known as the proliferating cell nuclear antigen
Proliferating cell nuclear antigen (PCNA) is a DNA clamp that acts as a processivity factor for DNA polymerase δ in eukaryotic cells and is essential for replication. PCNA is a homotrimer and achieves its processivity by encircling the DNA, whe ...
(PCNA). The PCNA ring has polarity with surfaces that interact with DNA polymerases and tethers them securely to the DNA template. PCNA-dependent stabilization of DNA polymerases has a significant effect on DNA replication because PCNAs are able to enhance the polymerase processivity up to 1,000-fold. PCNA is an essential cofactor and has the distinction of being one of the most common interaction platforms in the replisome to accommodate multiple processes at the replication fork, and so PCNA is also viewed as a regulatory cofactor for DNA polymerases.
Replication factor C
PCNA fully encircles the DNA template strand and must be loaded onto DNA at the replication fork. At the leading strand, loading of the PCNA is an infrequent process, because DNA replication on the leading strand is continuous until replication is terminated. However, at the lagging strand, DNA polymerase δ needs to be continually loaded at the start of each Okazaki fragment. This constant initiation of Okazaki fragment synthesis requires repeated PCNA loading for efficient DNA replication. PCNA loading is accomplished by thereplication factor C
The replication factor C, or RFC, is a five-subunit protein complex that is required for DNA replication.
The subunits of this heteropentamer are named Rfc1, Rfc2, Rfc3, Rfc4, and Rfc5 in ''Saccharomyces cerevisiae''. RFC is used in eukaryotic re ...
(RFC) complex. The RFC complex is composed of five ATPases: Rfc1, Rfc2, Rfc3, Rfc4 and Rfc5. RFC recognizes primer-template junctions and loads PCNA at these sites. The PCNA homotrimer is opened by RFC by ATP hydrolysis and is then loaded onto DNA in the proper orientation to facilitate its association with the polymerase. Clamp loaders can also unload PCNA from DNA; a mechanism needed when replication must be terminated.
Stalled replication fork
DNA replication
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all living organisms acting as the most essential part for biological inheritanc ...
at the replication fork can be halted by a shortage of deoxynucleotide triphosphates (dNTPs) or by DNA damage, resulting in replication stress
DNA replication stress refers to the state of a cell whose genome is exposed to various stresses. The events that contribute to replication stress occur during DNA replication, and can result in a stalled replication fork.
There are many events t ...
. This halting of replication is described as a stalled replication fork. A fork protection complex of proteins stabilizes the replication fork until DNA damage or other replication problems can be fixed. Prolonged replication fork stalling can lead to further DNA damage. Stalling signals are deactivated if the problems causing the replication fork are resolved.
Termination
 Termination of eukaryotic DNA replication requires different processes depending on whether the chromosomes are circular or linear. Unlike linear molecules, circular chromosomes are able to replicate the entire molecule. However, the two DNA molecules will remain linked together. This issue is handled by decatenation of the two DNA molecules by a
Termination of eukaryotic DNA replication requires different processes depending on whether the chromosomes are circular or linear. Unlike linear molecules, circular chromosomes are able to replicate the entire molecule. However, the two DNA molecules will remain linked together. This issue is handled by decatenation of the two DNA molecules by a type II topoisomerase
Type II topoisomerases are topoisomerases that cut both strands of the DNA helix simultaneously in order to manage DNA tangles and supercoils. They use the hydrolysis of ATP, unlike Type I topoisomerase. In this process, these enzymes change th ...
. Type II topoisomerases are also used to separate linear strands as they are intricately folded into a nucleosome within the cell.
As previously mentioned, linear chromosomes face another issue that is not seen in circular DNA replication. Due to the fact that an RNA primer is required for initiation of DNA synthesis, the lagging strand is at a disadvantage in replicating the entire chromosome. While the leading strand can use a single RNA primer to extend the 5' terminus of the replicating DNA strand, multiple RNA primers are responsible for lagging strand synthesis, creating Okazaki fragments. This leads to an issue due to the fact that DNA polymerase is only able to add to the 3' end of the DNA strand. The 3'-5' action of DNA polymerase along the parent strand leaves a short single-stranded DNA (ssDNA) region at the 3' end of the parent strand when the Okazaki fragments have been repaired. Since replication occurs in opposite directions at opposite ends of parent chromosomes, each strand is a lagging strand at one end. Over time this would result in progressive shortening of both daughter chromosomes. This is known as the end replication problem.
The end replication problem is handled in eukaryotic cells by telomere
A telomere (; ) is a region of repetitive nucleotide sequences associated with specialized proteins at the ends of linear chromosomes. Although there are different architectures, telomeres, in a broad sense, are a widespread genetic feature mos ...
regions and telomerase
Telomerase, also called terminal transferase, is a ribonucleoprotein that adds a species-dependent telomere repeat sequence to the 3' end of telomeres. A telomere is a region of repetitive sequences at each end of the chromosomes of most euka ...
. Telomeres extend the 3' end of the parental chromosome beyond the 5' end of the daughter strand. This single-stranded DNA structure can act as an origin of replication that recruits telomerase. Telomerase is a specialized DNA polymerase that consists of multiple protein subunits and an RNA component. The RNA component of telomerase anneals to the single stranded 3' end of the template DNA and contains 1.5 copies of the telomeric sequence. Telomerase contains a protein subunit that is a reverse transcriptase
A reverse transcriptase (RT) is an enzyme used to generate complementary DNA (cDNA) from an RNA template, a process termed reverse transcription. Reverse transcriptases are used by viruses such as HIV and hepatitis B to replicate their genomes, ...
called telomerase reverse transcriptase
Telomerase reverse transcriptase (abbreviated to TERT, or hTERT in humans) is a catalytic subunit of the enzyme telomerase, which, together with the telomerase RNA component (TERC), comprises the most important unit of the telomerase complex.
T ...
or TERT. TERT synthesizes DNA until the end of the template telomerase RNA and then disengages. This process can be repeated as many times as needed with the extension of the 3' end of the parental DNA molecule. This 3' addition provides a template for extension of the 5' end of the daughter strand by lagging strand DNA synthesis. Regulation of telomerase activity is handled by telomere-binding proteins.
Replication fork barriers
Prokaryotic DNA replication is bidirectional; within a replicative origin, replisome complexes are created at each end of the replication origin and replisomes move away from each other from the initial starting point. In prokaryotes, bidirectional replication initiates at one replicative origin on the circular chromosome and terminates at a site opposed from the initial start of the origin. These termination regions have DNA sequences known as ''Ter'' sites. These ''Ter'' sites are bound by the Tus protein. The ''Ter''-Tus complex is able to stop helicase activity, terminating replication. In eukaryotic cells, termination of replication usually occurs through the collision of the two replicative forks between two active replication origins. The location of the collision varies on the timing of origin firing. In this way, if a replication fork becomes stalled or collapses at a certain site, replication of the site can be rescued when a replisome traveling in the opposite direction completes copying the region. There are programmed replication fork barriers (RFBs) bound by RFB proteins in various locations, throughout the genome, which are able to terminate or pause replication forks, stopping progression of the replisome.Replication factories
It has been found that replication happens in a localised way in the cell nucleus. Contrary to the traditional view of moving replication forks along stagnant DNA, a concept of ''replication factories'' emerged, which means replication forks are concentrated towards some immobilised 'factory' regions through which the template DNA strands pass like conveyor belts.Replication factories , PavelHozák, Peter R.Cook, Trends in Cell Biology Volume 4 issue 2Sciencedirect
/ref>
Cell cycle regulation
cell cycle
The cell cycle, or cell-division cycle, is the series of events that take place in a cell that cause it to divide into two daughter cells. These events include the duplication of its DNA (DNA replication) and some of its organelles, and subs ...
. Progress through the cell cycle and in turn DNA replication is tightly regulated by the formation and activation of pre-replicative complexes (pre-RCs) which is achieved through the activation and inactivation of cyclin-dependent kinase
Cyclin-dependent kinases (CDKs) are the families of protein kinases first discovered for their role in regulating the cell cycle. They are also involved in regulating transcription, mRNA processing, and the differentiation of nerve cells. They a ...
s (Cdks, CDKs). Specifically it is the interactions of cyclin
Cyclin is a family of proteins that controls the progression of a cell through the cell cycle by activating cyclin-dependent kinase (CDK) enzymes or group of enzymes required for synthesis of cell cycle.
Etymology
Cyclins were originally disco ...
s and cyclin dependent kinases that are responsible for the transition from G1 into S-phase.
During the G1 phase of the cell cycle there are low levels of CDK activity. This low level of CDK activity allows for the formation of new pre-RC complexes but is not sufficient for DNA replication to be initiated by the newly formed pre-RCs. During the remaining phases of the cell cycle there are elevated levels of CDK activity. This high level of CDK activity is responsible for initiating DNA replication as well as inhibiting new pre-RC complex formation. Once DNA replication has been initiated the pre-RC complex is broken down. Due to the fact that CDK levels remain high during the S phase, G2, and M phases of the cell cycle no new pre-RC complexes can be formed. This all helps to ensure that no initiation can occur until the cell division is complete.
In addition to cyclin dependent kinases a new round of replication is thought to be prevented through the downregulation of Cdt1. This is achieved via degradation of Cdt1 as well as through the inhibitory actions of a protein known as geminin
Geminin, DNA replication inhibitor, also known as GMNN, is a protein in humans encoded by the ''GMNN'' gene. A nuclear protein present in most eukaryotes and highly conserved across species, numerous functions have been elucidated for geminin inc ...
. Geminin binds tightly to Cdt1 and is thought to be the major inhibitor of re-replication. Geminin first appears in S-phase and is degraded at the metaphase-anaphase transition, possibly through ubiquination by anaphase promoting complex
Anaphase-promoting complex (also called the cyclosome or APC/C) is an E3 ubiquitin ligase that marks target cell cycle proteins for degradation by the 26S proteasome. The APC/C is a large complex of 11–13 subunit proteins, including a cullin ...
(APC).
Various cell cycle checkpoint
Cell cycle checkpoints are control mechanisms in the eukaryotic cell cycle which ensure its proper progression. Each checkpoint serves as a potential termination point along the cell cycle, during which the conditions of the cell are assessed, wi ...
s are present throughout the course of the cell cycle that determine whether a cell will progress through division entirely. Importantly in replication the G1, or restriction, checkpoint makes the determination of whether or not initiation of replication will begin or whether the cell will be placed in a resting stage known as G0. Cells in the G0 stage of the cell cycle are prevented from initiating a round of replication because the minichromosome maintenance proteins are not expressed. Transition into the S-phase indicates replication has begun.
Replication checkpoint proteins
In order to preserve genetic information during cell division, DNA replication must be completed with high fidelity. In order to achieve this task, eukaryotic cells have proteins in place during certain points in the replication process that are able to detect any errors during DNA replication and are able to preserve genomic integrity. These checkpoint proteins are able to stop the cell cycle from entering mitosis in order to allow time for DNA repair. Checkpoint proteins are also involved in some DNA repair pathways, while they stabilize the structure of the replication fork to prevent further damage. These checkpoint proteins are essential to avoid passing down mutations or other chromosomal aberrations to offspring. Eukaryotic checkpoint proteins are well conserved and involve twophosphatidylinositol 3-kinase-related kinase
Phosphatidylinositol 3-kinase-related kinases (PIKKs) are a family of Ser/Thr-protein kinases with sequence similarity to phosphatidylinositol-3 kinases ( PI3Ks).
Members
The human PIKK family includes six members:
Structure
PIKKs protei ...
s (PIKKs), ATR ATR may refer to:
Medicine
* Acute transfusion reaction
* Ataxia telangiectasia and Rad3 related, a protein involved in DNA damage repair
Science and mathematics
* Advanced Test Reactor, nuclear research reactor at the Idaho National Laboratory, ...
and ATM. Both ATR and ATM share a target phosphorylation sequence, the SQ/TQ motif, but their individual roles in cells differ.
ATR is involved in arresting the cell cycle in response to DNA double-stranded breaks. ATR has an obligate checkpoint partner, ATR-interacting-protein (ATRIP), and together these two proteins are responsive to stretches of single-stranded DNA that are coated by replication protein A (RPA). The formation of single-stranded DNA occurs frequently, more often during replication stress. ATR-ATRIP is able to arrest the cell cycle to preserve genome integrity. ATR is found on chromatin during S phase, similar to RPA and claspin.
The generation of single-stranded DNA tracts is important in initiating the checkpoint pathways downstream of replication damage. Once single-stranded DNA becomes sufficiently long, single-stranded DNA coated with RPA are able to recruit ATR-ATRIP. In order to become fully active, the ATR kinase rely on sensor proteins that sense whether the checkpoint proteins are localized to a valid site of DNA replication stress. The RAD9-HUS1
Checkpoint protein HUS1 is a protein that in humans is encoded by the ''HUS1'' gene.
Function
The protein encoded by this gene is a component of an evolutionarily conserved, genotoxin-activated checkpoint complex that is involved in the cell cy ...
- Rad1 (9-1-1) heterotrimeric clamp and its clamp loader RFCRad17 are able to recognize gapped or nicked DNA. The RFCRad17 clamp loader loads 9-1-1 onto the damaged DNA. The presence of 9-1-1 on DNA is enough to facilitate the interaction between ATR-ATRIP and a group of proteins termed checkpoint mediators, such as TOPBP1
DNA topoisomerase 2-binding protein 1 (TOPBP1) is a scaffold protein that in humans is encoded by the ''TOPBP1'' gene.
TOPBP1 was first identified as a protein binding partner of DNA topoisomerase-IIβ by a yeast 2-hybrid screen, giving it its ...
and Mrc1/ claspin. TOPBP1 interacts with and recruits the phosphorylated Rad9 component of 9-1-1 and binds ATR-ATRIP, which phosphorylates Chk1. Mrc1/Claspin is also required for
the complete activation of ATR-ATRIP that phosphorylates Chk1, the major downstream checkpoint effector kinase. Claspin is a component of the replisome and contains a domain for docking with Chk1, revealing a specific function of Claspin during DNA replication: the promotion of
checkpoint signaling at the replisome.
Chk1
Checkpoint kinase 1, commonly referred to as Chk1, is a serine/threonine-specific protein kinase that, in humans, is encoded by the ''CHEK1'' gene. Chk1 coordinates the DNA damage response (DDR) and cell cycle checkpoint response. Activation of Chk ...
signaling is vital for arresting the cell cycle and preventing cells from entering mitosis with incomplete DNA replication or DNA damage. The Chk1-dependent Cdk inhibition is important for the function of the ATR-Chk1 checkpoint and to arrest the cell cycle and allow sufficient time for completion of DNA repair mechanisms, which in turn prevents the inheritance of damaged DNA. In addition, Chk1-dependent Cdk inhibition plays a critical role in inhibiting origin firing during S phase. This mechanism prevents continued DNA synthesis and is required for the protection of the genome in the
presence of replication stress and potential genotoxic conditions. Thus, ATR-Chk1 activity further prevents potential replication problems at the level of single replication origins by inhibiting initiation of replication throughout the genome, until the signaling cascade maintaining cell-cycle arrest is turned off.
Replication through nucleosomes
histone
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei. They act as spools around which DNA winds to create structural units called nucleosomes. Nucleosomes in turn are wr ...
proteins, forming a nucleosome
A nucleosome is the basic structural unit of DNA packaging in eukaryotes. The structure of a nucleosome consists of a segment of DNA wound around eight histone proteins and resembles thread wrapped around a spool. The nucleosome is the fundamen ...
. The nucleosome octamer includes two copies of each histone H2A, H2B, H3, and H4. Due to the tight association of histone proteins to DNA, eukaryotic cells have proteins that are designed to remodel histones ahead of the replication fork, in order to allow smooth progression of the replisome. There are also proteins involved in reassembling histones behind the replication fork to reestablish the nucleosome conformation.
There are several histone chaperones that are known to be involved in nucleosome assembly after replication. The FACT
A fact is a datum about one or more aspects of a circumstance, which, if accepted as true and proven true, allows a logical conclusion to be reached on a true–false evaluation. Standard reference works are often used to check facts. Scient ...
complex has been found to interact with DNA polymerase α-primase complex, and the subunits of the FACT complex interacted genetically with replication factors. The FACT complex is a heterodimer that does not hydrolyze ATP, but is able to facilitate "loosening" of histones in nucleosomes, but how the FACT complex is able to relieve the tight association of histones for DNA removal remains unanswered.
Another histone chaperone that associates with the replisome is Asf1, which interacts with the Mcm complex dependent on histone dimers H3-H4. Asf1 is able to pass newly synthesized H3-H4 dimer to deposition factors behind the replication fork and this activity makes the H3-H4 histone dimers available at the site of histone deposition just after replication. Asf1 (and its partner Rtt109) has also been implicated in inhibiting gene expression from replicated genes during S-phase.
The heterotrimeric chaperone chromatin assembly factor 1 (CAF-1
Chromatin assembly factor-1 (CAF-1) is a protein complex — including Chaf1a (p150), Chaf1b (p60), and p48 subunits in humans, or Cac1, Cac2, and Cac3, respectively, in yeast— that assembles histone tetramers onto replicating DNA during the ...
) is a chromatin formation protein that is involved in depositing histones onto both newly replicated DNA strands to form chromatin. CAF-1 contains a PCNA-binding motif, called a PIP-box, that allows CAF-1 to associate with the replisome through PCNA and is able to deposit histone H3-H4 dimers onto newly synthesized DNA. The Rtt106 chaperone is also involved in this process, and associated with CAF-1 and H3-H4 dimers during chromatin formation. These processes load newly synthesized histones onto DNA.
After the deposition of histones H3-H4, nucleosomes form by the association of histone H2A-H2B. This process is thought to occur through the FACT complex, since it already associated with the replisome and is able to bind free H2A-H2B, or there is the possibility of another H2A-H2B chaperone, Nap1. Electron microscopy studies show that this occurs very quickly, as nucleosomes can be observed forming just a few hundred base pairs after the replication fork. Therefore, the entire process of forming new
nucleosomes takes place just after replication due to the coupling of histone chaperones to the replisome.
Comparisons between prokaryotic and eukaryotic DNA replication
When compared toprokaryotic DNA replication
Prokaryotic DNA Replication is the process by which a prokaryote duplicates its DNA into another copy that is passed on to daughter cells. Although it is often studied in the model organism '' E. coli'', other bacteria show many similarities. ...
, the completion of eukaryotic DNA replication is more complex and involves multiple origins of replication
The origin of replication (also called the replication origin) is a particular sequence in a genome at which replication is initiated. Propagation of the genetic material between generations requires timely and accurate duplication of DNA by semi ...
and replicative proteins to accomplish. Prokaryotic DNA is arranged in a circular shape, and has only one replication origin when replication starts. By contrast, eukaryotic DNA is linear. When replicated, there are as many as one thousand origins of replication.
Eukaryotic DNA is bidirectional. Here the meaning of the word bidirectional is different. Eukaryotic linear DNA has many origins (called O) and termini (called T). "T" is present to the right of "O". One "O" and one "T" together form one replicon. After the formation of pre-initiation complex, when one replicon starts elongation, initiation starts in second replicon. Now, if the first replicon moves in clockwise direction, the second replicon moves in anticlockwise direction, until "T" of first replicon is reached. At "T", both the replicons merge to complete the process of replication. Meanwhile, the second replicon is moving in forward direction also, to meet with the third replicon. This clockwise and counter-clockwise movement of two replicons is termed as bidirectional replication.
Eukaryotic DNA replication requires precise coordination of all DNA polymerases and associated proteins to replicate the entire genome each time a cell divides. This process is achieved through a series of steps of protein assemblies at origins of replication, mainly focusing the regulation of DNA replication on the association of the MCM helicase with the DNA. These origins of replication direct the number of protein complexes that will form to initiate replication. In prokaryotic DNA replication regulation focuses on the binding of the DnaA initiator protein to the DNA, with initiation of replication occurring multiple times during one cell cycle. Both prokaryotic and eukaryotic DNA use ATP binding and hydrolysis to direct helicase loading and in both cases the helicase is loaded in the inactive form. However, eukaryotic helicases are double hexamers that are loaded onto double stranded DNA whereas prokaryotic helicases are single hexamers loaded onto single stranded DNA.
Segregation of chromosomes is another difference between prokaryotic and eukaryotic cells. Rapidly dividing cells, such as bacteria, will often begin to segregate chromosomes that are still in the process of replication. In eukaryotic cells chromosome segregation into the daughter cells is not initiated until replication is complete in all chromosomes. Despite these differences, however, the underlying process of replication is similar for both prokaryotic and eukaryotic DNA.
Eukaryotic DNA replication protein list
List of major proteins involved in eukaryotic DNA replication:See also
*DNA replication
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all living organisms acting as the most essential part for biological inheritanc ...
*Prokaryotic DNA replication
Prokaryotic DNA Replication is the process by which a prokaryote duplicates its DNA into another copy that is passed on to daughter cells. Although it is often studied in the model organism '' E. coli'', other bacteria show many similarities. ...
*Processivity
In molecular biology and biochemistry, processivity is an enzyme's ability to catalyze "consecutive reactions without releasing its substrate".
For example, processivity is the average number of nucleotides added by a polymerase enzyme, such as ...
References
{{DNA replication DNA replication