Diguanylate Cyclase on:
[Wikipedia]
[Google]
[Amazon]
In
 As of mid-2011, 11 crystal structures of confirmed or putative DGCs have been solved, with PDB accession codes , , , , , , , , , , and .
As of mid-2011, 11 crystal structures of confirmed or putative DGCs have been solved, with PDB accession codes , , , , , , , , , , and .

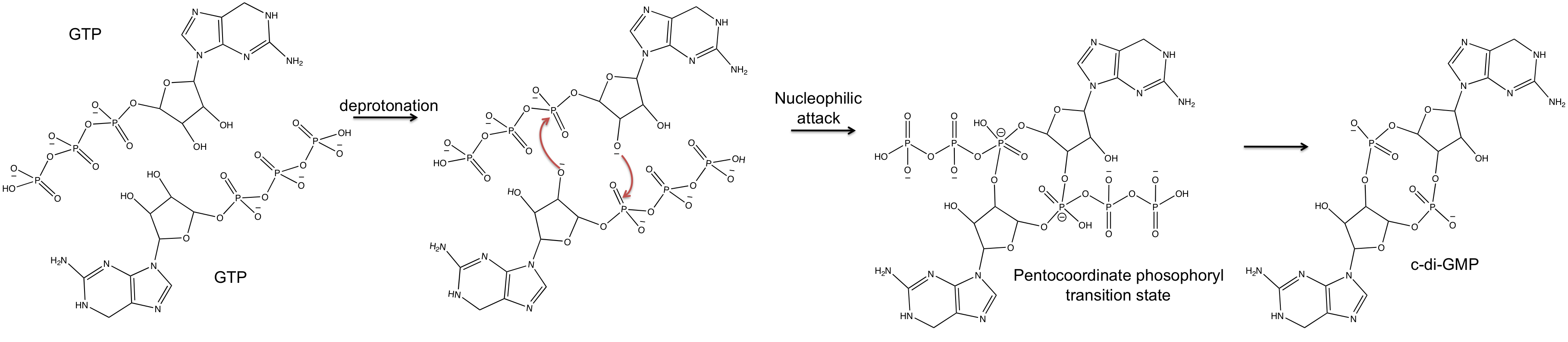
enzymology
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. A ...
, diguanylate cyclase, also known as diguanylate kinase (), is an enzyme that catalyzes
Catalysis () is the process of increasing the rate of a chemical reaction by adding a substance known as a catalyst (). Catalysts are not consumed in the reaction and remain unchanged after it. If the reaction is rapid and the catalyst recyc ...
the chemical reaction:
2 Guanosine triphosphate ↔ 2 diphosphate + cyclic di-3',5'-guanylate
The substrates of diguanylate cyclases (DGCs) are two molecules of guanosine triphosphate (GTP) and the products are two molecules of diphosphate and one molecule of cyclic di-3’,5’-guanylate (cyclic di-GMP
Cyclic di-GMP (also called cyclic diguanylate and c-di-GMP) is a second messenger used in signal transduction in a wide variety of bacteria. Cyclic di-GMP is not known to be used by archaea, and has only been observed in eukaryotes in ''Dictyostel ...
).
Degradation of cyclic di-GMP to guanosine monophosphate
Guanosine monophosphate (GMP), also known as 5′-guanidylic acid or guanylic acid (conjugate base guanylate), is a nucleotide that is used as a monomer in RNA. It is an ester of phosphoric acid with the nucleoside guanosine. GMP consists of the ...
(GMP) is catalyzed by a phosphodiesterase (PDE).
Structure
Diguanylate cyclases are characterized by the conserved amino acid sequence motifs “ GGDEF” (Gly
Glycine (symbol Gly or G; ) is an amino acid that has a single hydrogen atom as its side chain. It is the simplest stable amino acid (carbamic acid is unstable), with the chemical formula NH2‐ CH2‐ COOH. Glycine is one of the proteinogeni ...
-Gly-Asp
Asp may refer to:
Places
* Asp, part of Densbüren, Aargau, Switzerland
* Aspe (''Asp'' in Valencian), Alicante, Spain
* Asp Lake, a lake in Minnesota
Animals
* Asp (fish)
* Asp (snake), in antiquity, one of several venomous snakes
** ''Cera ...
- Glu- Phe) or “GGEEF” (Gly-Gly-Glu-Glu-Phe), which constitute the domain
Domain may refer to:
Mathematics
*Domain of a function, the set of input values for which the (total) function is defined
**Domain of definition of a partial function
**Natural domain of a partial function
**Domain of holomorphy of a function
* Do ...
of the DGC active site
In biology and biochemistry, the active site is the region of an enzyme where substrate molecules bind and undergo a chemical reaction. The active site consists of amino acid residues that form temporary bonds with the substrate (binding site) a ...
.
These domains are often found coupled to other signaling domains within multidomain proteins
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, respo ...
. Often, GGDEF domains with DGC activity are found in the same proteins as c-di-GMP-specific phosphodiesterase (PDE) EAL (Glu-Ala Ala, ALA, Alaa or Alae may refer to:
Places
* Ala, Hiiu County, Estonia, a village
* Ala, Valga County, Estonia, a village
* Ala, Alappuzha, Kerala, India, a village
* Ala, Iran, a village in Semnan Province
* Ala, Gotland, Sweden
* Alad, Seydu ...
- Leu) domains.
DGC is thought to only be active as a dimer
Dimer may refer to:
* Dimer (chemistry), a chemical structure formed from two similar sub-units
** Protein dimer, a protein quaternary structure
** d-dimer
* Dimer model, an item in statistical mechanics, based on ''domino tiling''
* Julius Dimer ( ...
consisting of two subunits, both with GGDEF domains.
The active (or catalytic) site is located at the interface between the two subunits, each binding one molecule of GTP. (See Activation mechanism and Regulation section for more information)
Weak sequence similarity and pronounced secondary structure
Protein secondary structure is the three dimensional conformational isomerism, form of ''local segments'' of proteins. The two most common Protein structure#Secondary structure, secondary structural elements are alpha helix, alpha helices and beta ...
similarity between GGDEF domains and the catalytic domains of adenylate cyclases (AC) have led to the hypothesis that DGCs and ACs share a similar fold.
This was verified with the resolution of the crystal structure of the DGC PleD from ''Caulobacter crescentus
''Caulobacter crescentus'' is a Gram-negative, oligotrophic bacterium widely distributed in fresh water lakes and streams. The taxon is more properly known as ''Caulobacter vibrioides'' (Henrici and Johnson 1935).
''C. crescentus'' is an importa ...
'' in complex with c-di-GMP. As shown in the figure, active PleD, shown as a dimer, is composed of the catalytic DCG domain (labeled DGC) and two CheY-like receiver domains (labeled D1/D2). The DGC domain of each subunit is linked to the two CheY-like domains by a flexible peptide linkage chain. The DCG domain closely resembles the domain of the AC catalytic core which consists of a five-stranded β-sheet surrounded by helices
A helix () is a shape like a corkscrew or spiral staircase. It is a type of smooth space curve with tangent lines at a constant angle to a fixed axis. Helices are important in biology, as the DNA molecule is formed as two intertwined helices, ...
.
 As of mid-2011, 11 crystal structures of confirmed or putative DGCs have been solved, with PDB accession codes , , , , , , , , , , and .
As of mid-2011, 11 crystal structures of confirmed or putative DGCs have been solved, with PDB accession codes , , , , , , , , , , and .
Biological function
Diguanylate cyclase participate in the formation of the ubiquitoussecond messenger
Second messengers are intracellular signaling molecules released by the cell in response to exposure to extracellular signaling molecules—the first messengers. (Intercellular signals, a non-local form or cell signaling, encompassing both first me ...
, cyclic-di-GMP, involved in bacterial biofilm formation and persistence. The GGDEF domain was first identified in the regulatory protein, PleD of the bacterium ''Caulobacter crescentus
''Caulobacter crescentus'' is a Gram-negative, oligotrophic bacterium widely distributed in fresh water lakes and streams. The taxon is more properly known as ''Caulobacter vibrioides'' (Henrici and Johnson 1935).
''C. crescentus'' is an importa ...
''.
It was later noted that numerous bacterial genomes encoded multiple proteins with a GGDEF domain.
'' Pseudomonas aeruginosa'' PAO1 has 33 proteins with GGDEF domains, '' Escherichia coli'' K-12 has 19, and '' Vibrio cholerae'' O1 has 41.
In the cell cycle of ''Caulobacter crescentus
''Caulobacter crescentus'' is a Gram-negative, oligotrophic bacterium widely distributed in fresh water lakes and streams. The taxon is more properly known as ''Caulobacter vibrioides'' (Henrici and Johnson 1935).
''C. crescentus'' is an importa ...
'', DGC PleD is known to control pole morphogenesis.
In ''Pseudomonas fluorescens
''Pseudomonas fluorescens'' is a common Gram-negative, rod-shaped bacterium. It belongs to the ''Pseudomonas'' genus; 16S rRNA analysis as well as phylogenomic analysis has placed ''P. fluorescens'' in the ''P. fluorescens'' group within the genu ...
'' DGC WspR activity is hypothesized to be partially responsible for the wrinkly spreader (WS) phenotype.
In '' Pseudomonas aeruginosa'', WspR has also been known to control autoaggregation.
''Role of DGC in C. crescentus cell cycle''
During the cell cycle of ''C. crescentus'', proteins with GGDEF and EAL domains are separated towards the two distinct poles. The active form of diguanylate cyclase PleD localizes to the stalked pole of differentiating ''C. crescentus'' cells. It has been suggested that the function of PleD is two-fold. PleD is responsible for turning off flagellum rotations and inhibiting motility before genome replication begins and also for regenerating motility after differentiation has completed.
Activation Mechanism and Regulation
The crystal structure of the ''C. crescentus'' diguanylate cyclase, PleD, contains three domains; a GGDEF domain with diguanylate cyclase activity and two CheY-like receiver domains (D1/D2). As seen in the figure, the active form of PleD is a dimer which forms by phosphorylation of the first receiver domain (D1). Phosphorylation of the receiver domain increases the dimerization affinity by approximately 10-fold over non-phosphorylated domains. Inhibition of DGC activity is thought to beallosteric
In biochemistry, allosteric regulation (or allosteric control) is the regulation of an enzyme by binding an effector molecule at a site other than the enzyme's active site.
The site to which the effector binds is termed the ''allosteric site ...
and non-competitive.
Cyclic di-GMP binds to interface between the DGC and D2 domains stabilizing the open structure and preventing catalysis.
Strong product inhibition has been observed with a Ki of 0.5 μM.
Though the exact catalytic mechanism has not been resolved, it is hypothesized that the dimerized structure of PleD facilitates interaction of the two GTP molecules within the DGC active site for cyclization. A proposed mechanism by Chan et al. indicates that the 3'- OH group of the GTP is deprotonated by a glutamic acid residue (E370) to allow for intermolecular nucleophilic attack of the α- phosphate. The pentachoordinated transition state created through this nucleophilic attack is possibly stabilized by a Lysine residue (K332).
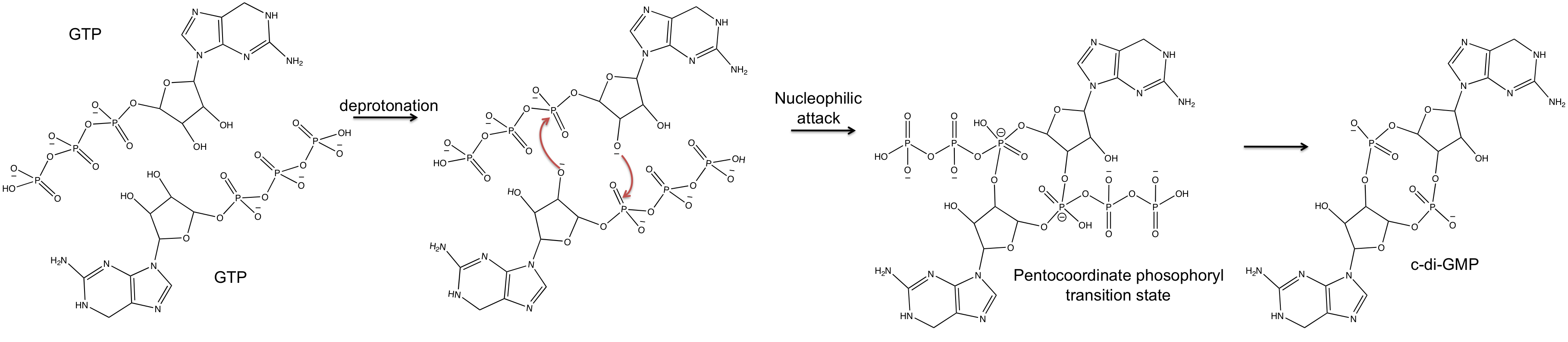
References
Further reading
* * * {{DEFAULTSORT:Diguanylate Cyclase EC 2.7.7