Decapping complex on:
[Wikipedia]
[Google]
[Amazon]
 The
The

 The
The mRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of Protein biosynthesis, synthesizing a protein.
mRNA is ...
decapping complex is a protein complex in eukaryotic cells responsible for removal of the 5' cap
In molecular biology, the five-prime cap (5′ cap) is a specially altered nucleotide on the 5′ end of some primary transcripts such as precursor messenger RNA. This process, known as mRNA capping, is highly regulated and vital in the creation ...
. The active enzyme of the decapping complex is the bilobed Nudix family enzyme Dcp2
mRNA-decapping enzyme 2 is a protein that in humans is encoded by the ''DCP2'' gene
In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleotides in DNA that is ...
, which hydrolyzes 5' cap
In molecular biology, the five-prime cap (5′ cap) is a specially altered nucleotide on the 5′ end of some primary transcripts such as precursor messenger RNA. This process, known as mRNA capping, is highly regulated and vital in the creation ...
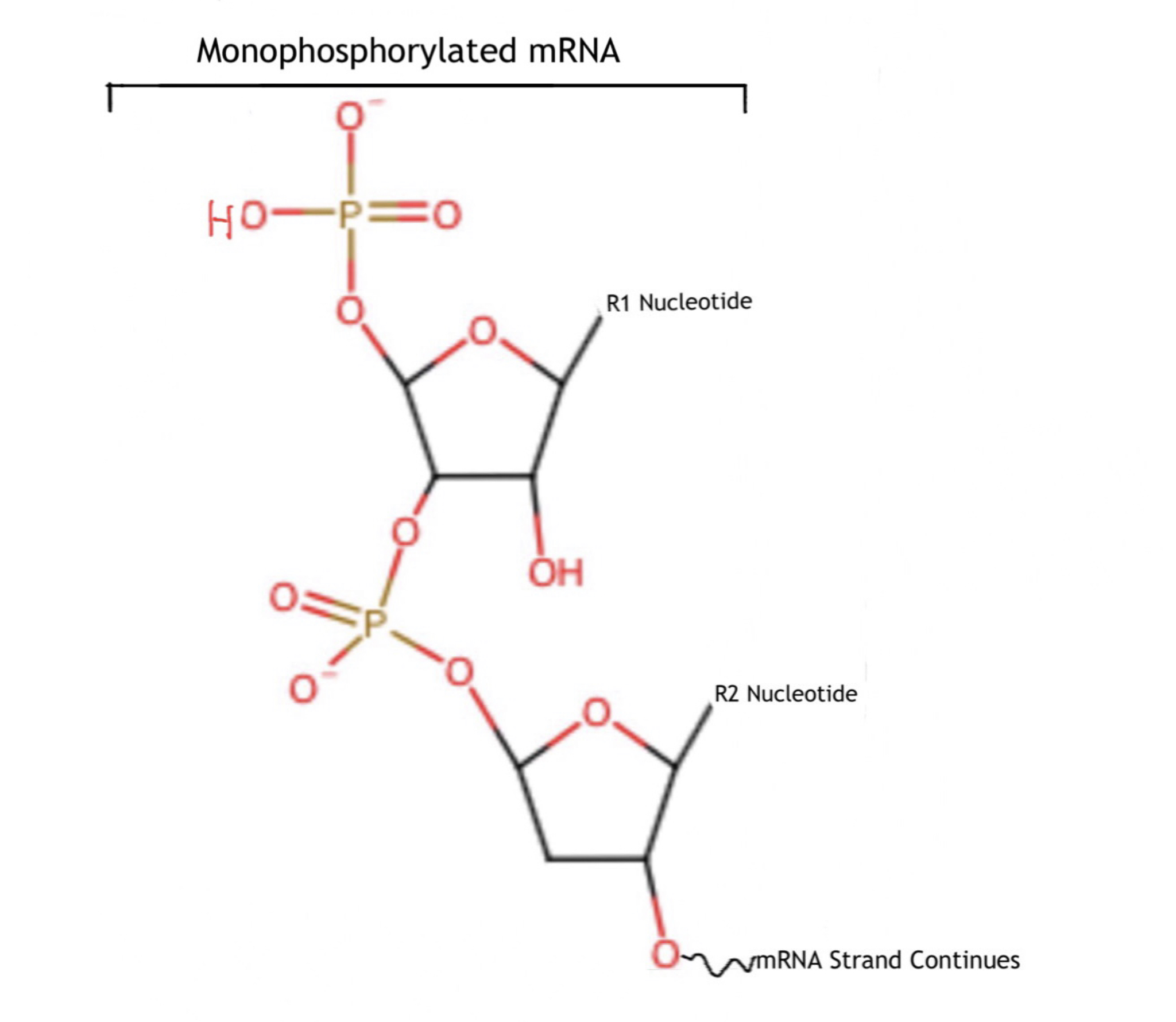
and releases 7mGDP and a 5'-monophosphorylated mRNA. This decapped mRNA is inhibited for translation and will be degraded by exonuclease
Exonucleases are enzymes that work by cleaving nucleotides one at a time from the end (exo) of a polynucleotide chain. A hydrolyzing reaction that breaks phosphodiester bonds at either the 3′ or the 5′ end occurs. Its close relative is th ...
s. The core decapping complex is conserved in eukaryotes. Dcp2 is activated by Decapping Protein 1 (Dcp1) and in higher eukaryote
The eukaryotes ( ) constitute the Domain (biology), domain of Eukaryota or Eukarya, organisms whose Cell (biology), cells have a membrane-bound cell nucleus, nucleus. All animals, plants, Fungus, fungi, seaweeds, and many unicellular organisms ...
s joined by the scaffold protein VCS. Together with many other accessory proteins, the decapping complex assembles in P-bodies
In cellular biology, P-bodies, or processing bodies, are distinct foci formed by phase separation within the cytoplasm of a eukaryotic cell consisting of many enzymes involved in mRNA turnover. P-bodies are highly conserved structures and have ...
in the cytoplasm
The cytoplasm describes all the material within a eukaryotic or prokaryotic cell, enclosed by the cell membrane, including the organelles and excluding the nucleus in eukaryotic cells. The material inside the nucleus of a eukaryotic cell a ...
.
Purpose of the decapping complex
mRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of Protein biosynthesis, synthesizing a protein.
mRNA is ...
needs to be degraded, or else it will keep floating around the cell and create unwanted proteins at random. The mRNA 5' cap is specifically designed to keep mRNA from being degraded before it can be used, and so needs to be removed so the mRNA decay pathway can take care of it.
Decapping mechanism
Dcp2
mRNA-decapping enzyme 2 is a protein that in humans is encoded by the ''DCP2'' gene
In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleotides in DNA that is ...
is the protein that actually decaps mRNA, and the rest of proteins in the complex enhance its function and allow it to hydrolyze
Hydrolysis (; ) is any chemical reaction in which a molecule of water breaks one or more chemical bonds. The term is used broadly for substitution, elimination, and solvation reactions in which water is the nucleophile.
Biological hydrolysis ...
the chemical bond attaching the mRNA to the 5' cap
In molecular biology, the five-prime cap (5′ cap) is a specially altered nucleotide on the 5′ end of some primary transcripts such as precursor messenger RNA. This process, known as mRNA capping, is highly regulated and vital in the creation ...
. The Nudix domain in Dcp2 hydrolyzes
Hydrolysis (; ) is any chemical reaction in which a molecule of water breaks one or more chemical bonds. The term is used broadly for substitution reaction, substitution, elimination reaction, elimination, and solvation reactions in which water ...
one of the bonds on the triphosphate bridge that hooks the mRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of Protein biosynthesis, synthesizing a protein.
mRNA is ...
and the 5' cap
In molecular biology, the five-prime cap (5′ cap) is a specially altered nucleotide on the 5′ end of some primary transcripts such as precursor messenger RNA. This process, known as mRNA capping, is highly regulated and vital in the creation ...
together, causing the 7-methylguanosine
7-Methylguanosine (m7G) is a modified purine nucleoside. It is a methylated version of guanosine and when found in human urine, it may be a biomarker of some types of cancer. In the RNAs, 7-methylguanosine have been used to study and examine t ...
cap to come off and leaving the mRNA open to degradation by the exonuclease
Exonucleases are enzymes that work by cleaving nucleotides one at a time from the end (exo) of a polynucleotide chain. A hydrolyzing reaction that breaks phosphodiester bonds at either the 3′ or the 5′ end occurs. Its close relative is th ...
s in the cell.
Structure of the decapping complex
Both single-celled and multicellular organisms need to decap their mRNA to get rid of it, but different organisms have slightly different proteins that carry out this process. There are many proteins that stay the same, but several key differences between the single-celled (yeast
Yeasts are eukaryotic, single-celled microorganisms classified as members of the fungus kingdom (biology), kingdom. The first yeast originated hundreds of millions of years ago, and at least 1,500 species are currently recognized. They are est ...
) and multicellular (metazoan
Animals are multicellular, eukaryotic organisms in the biological kingdom Animalia (). With few exceptions, animals consume organic material, breathe oxygen, have myocytes and are able to move, can reproduce sexually, and grow from a ho ...
) decapping complexes.
Yeast decapping complex
In yeast (''S. cerevisiae
''Saccharomyces cerevisiae'' () (brewer's yeast or baker's yeast) is a species of yeast (single-celled fungal microorganisms). The species has been instrumental in winemaking, baking, and brewing since ancient times. It is believed to have been o ...
''), Dcp2 is joined by the decapping activator Dcp1, the helicase
Helicases are a class of enzymes that are vital to all organisms. Their main function is to unpack an organism's genetic material. Helicases are motor proteins that move directionally along a nucleic double helix, separating the two hybridized ...
Dhh1, the exonuclease
Exonucleases are enzymes that work by cleaving nucleotides one at a time from the end (exo) of a polynucleotide chain. A hydrolyzing reaction that breaks phosphodiester bonds at either the 3′ or the 5′ end occurs. Its close relative is th ...
Xrn1, nonsense mediated decay
Nonsense-mediated mRNA decay (NMD) is a surveillance pathway that exists in all eukaryotes. Its main function is to reduce errors in gene expression by eliminating mRNA transcripts that contain premature stop codons. Translation of these aberran ...
factors Upf1, Upf2, and Upf3, the LSm
LSM may refer to:
Science
*Laboratoire Souterrain de Modane (Modane Underground Laboratory), a particle physics laboratory in France
*Lanthanum strontium manganite, a crystal used as a cathode material
*Confocal microscopy, Laser scanning microsc ...
complex, Pat1, and various other proteins. These proteins all localize to cytoplasmic structures called P-bodies
In cellular biology, P-bodies, or processing bodies, are distinct foci formed by phase separation within the cytoplasm of a eukaryotic cell consisting of many enzymes involved in mRNA turnover. P-bodies are highly conserved structures and have ...
. Notably in yeast there are no translation factors or ribosomal proteins inside P-bodies.
Metazoan decapping complex
Higher eukaryotes have slightly different members of the decapping complex. The enzyme Dcp2 is still the catalytic subunit which forms aholoenzyme
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different mol ...
with Dcp1, and interacts with auxiliary proteins such as Xrn1
5′-3′ exoribonuclease 1 (Xrn1) is a protein that in humans is encoded by the XRN1 gene. Xrn1 hydrolyses RNA in the 5′ to 3′ direction.
Function
This gene encodes a member of the 5′-3′ exonuclease family. The encoded protein may be ...
, Upf1
Regulator of nonsense transcripts 1 (or Up-frameshift suppressor 1 homolog) is a protein that in humans is encoded by the ''UPF1'' gene.
Function
This gene encodes a protein that is part of a post-splicing multiprotein complex, the exon junct ...
, Upf2
Regulator of nonsense transcripts 2 is a protein that in humans is encoded by the ''UPF2'' gene.
Function
This gene encodes a protein that is part of a post-splicing multiprotein complex, the exon junction complex, involved in both mRNA nucle ...
, Upf3, the LSm complex, and the Dhh1 ortholog DDX6. Proteins unique to plants and mammals include the beta propeller
In structural biology, a beta-propeller (β-propeller) is a type of all-β protein architecture characterized by 4 to 8 highly symmetrical blade-shaped beta sheets arranged toroidally around a central axis. Together the beta-sheets form a funnel- ...
protein Hedls and the enhancer of decapping Edc3. Researchers know how the complex physically associates because of immunoprecipitation
Immunoprecipitation (IP) is the technique of precipitating a protein antigen out of solution using an antibody that specifically binds to that particular protein. This process can be used to isolate and concentrate a particular protein from a sam ...
, while structural details of each part of the complex have been discovered by using x-ray crystallography
X-ray crystallography is the experimental science of determining the atomic and molecular structure of a crystal, in which the crystalline structure causes a beam of incident X-rays to Diffraction, diffract in specific directions. By measuring th ...
in conjunction with protein crystallization
Protein crystallization is the process of formation of a regular array of individual protein molecules stabilized by crystal contacts. If the crystal is sufficiently ordered, it will diffract. Some proteins naturally form crystalline arrays, ...
. Each of these proteins contribute different things to the decapping complex, as discussed below.
Dcp2

Dcp2
mRNA-decapping enzyme 2 is a protein that in humans is encoded by the ''DCP2'' gene
In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleotides in DNA that is ...
, as the main catalyst
Catalysis () is the increase in rate of a chemical reaction due to an added substance known as a catalyst (). Catalysts are not consumed by the reaction and remain unchanged after it. If the reaction is rapid and the catalyst recycles quick ...
of the decapping process, relies on a specific pattern of amino acids called a nudix domain to align itself with the 5' cap
In molecular biology, the five-prime cap (5′ cap) is a specially altered nucleotide on the 5′ end of some primary transcripts such as precursor messenger RNA. This process, known as mRNA capping, is highly regulated and vital in the creation ...
in order to hydrolyze
Hydrolysis (; ) is any chemical reaction in which a molecule of water breaks one or more chemical bonds. The term is used broadly for substitution, elimination, and solvation reactions in which water is the nucleophile.
Biological hydrolysis ...
it. A nudix domain
A domain is a geographic area controlled by a single person or organization. Domain may also refer to:
Law and human geography
* Demesne, in English common law and other Medieval European contexts, lands directly managed by their holder rather ...
is made by packing two beta sheet
The beta sheet (β-sheet, also β-pleated sheet) is a common motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally by at least two or three backbone hydrogen bonds, forming a gene ...
s between multiple alpha helices
An alpha helix (or α-helix) is a sequence of amino acids in a protein that are twisted into a coil (a helix).
The alpha helix is the most common structural arrangement in the secondary structure of proteins. It is also the most extreme type of l ...
, can be various lengths and sizes, and is generally used by proteins to carry out dephosphorylation
In biochemistry, dephosphorylation is the removal of a phosphate () group from an organic compound by hydrolysis. It is a reversible post-translational modification. Dephosphorylation and its counterpart, phosphorylation, activate and deactivate e ...
, getting rid of a phosphate
Phosphates are the naturally occurring form of the element phosphorus.
In chemistry, a phosphate is an anion, salt, functional group or ester derived from a phosphoric acid. It most commonly means orthophosphate, a derivative of orthop ...
by inserting a water molecule into the bond between the phosphate and the rest of the molecule. In the case of Dcp2, it contains multiple glutamic acid
Glutamic acid (symbol Glu or E; known as glutamate in its anionic form) is an α- amino acid that is used by almost all living beings in the biosynthesis of proteins. It is a non-essential nutrient for humans, meaning that the human body can ...
side chains that are negatively charged in normal cellular conditions, and these are what allow the protein to manipulate water molecules to hydrolyze the tri-phosphate bridge that connects the 5' end of the mRNA to the 7-methylguanosine
7-Methylguanosine (m7G) is a modified purine nucleoside. It is a methylated version of guanosine and when found in human urine, it may be a biomarker of some types of cancer. In the RNAs, 7-methylguanosine have been used to study and examine t ...
cap. Therefore, the nudix domain is what allows Dcp2 to remove the 5' cap, which results in the creation 7mGDP, a 7-methylguanosine with two phosphate groups attached, and a monophosphorylated mRNA strand.
Before the nudix domain is an N-terminal regulatory domain (NRD), which further helps hydrolyze the 5' mRNA cap. After the nudix domain is a C-terminal area called Box B, which helps bind Dcp2 to RNA. With all three of these main motifs, Dcp2 is able to find, bind firmly to, and hydrolyzes a 5' mRNA cap. It does this either by recognizing a hairpin loop
Stem-loops are nucleic acid secondary structural elements which form via intramolecular base pairing in single-stranded DNA or RNA. They are also referred to as hairpins or hairpin loops. A stem-loop occurs when two regions of the same nucleic ...
in the RNA within 10 base pair
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA ...
s of the cap, which is called a Dcp2 binding and decapping element, or by a separate protein recognizing a base pair pattern in the mRNA and directly recruiting the Dcp2-Dcp1 holoenzyme. Unfortunately, Dcp2 works slowly, and needs a few other proteins to coordinate with it so it can decap mRNA in a timely manner.
Dcp1
Dcp1 is a regulatory subunit, it combines with Dcp2, creating aholoenzyme
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different mol ...
that can decap mRNA properly. Without Dcp1, it is actually impossible for Dcp2 to decap anything in vivo
Studies that are ''in vivo'' (Latin for "within the living"; often not italicized in English) are those in which the effects of various biological entities are tested on whole, living organisms or cells, usually animals, including humans, an ...
, and it only works incredibly slowly in vitro
''In vitro'' (meaning ''in glass'', or ''in the glass'') Research, studies are performed with Cell (biology), cells or biological molecules outside their normal biological context. Colloquially called "test-tube experiments", these studies in ...
, which makes forming this holoenzyme an essential process in decapping.
Dcp1's secondary structure
Protein secondary structure is the local spatial conformation of the polypeptide backbone excluding the side chains. The two most common Protein structure#Secondary structure, secondary structural elements are alpha helix, alpha helices and beta ...
consists of seven beta sheet
The beta sheet (β-sheet, also β-pleated sheet) is a common motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally by at least two or three backbone hydrogen bonds, forming a gene ...
s and three alpha helices
An alpha helix (or α-helix) is a sequence of amino acids in a protein that are twisted into a coil (a helix).
The alpha helix is the most common structural arrangement in the secondary structure of proteins. It is also the most extreme type of l ...
. which come together to form a V-shaped tertiary structure
Protein tertiary structure is the three-dimensional shape of a protein. The tertiary structure will have a single polypeptide chain "backbone" with one or more protein secondary structures, the protein domains. Amino acid side chains and the ...
. The defining features of Dcp1 are the EVH1 domain
WH1 domains, also known as EVH1 domains, are evolutionary conserved protein domain, protein domains found on WASP (VASP) proteins, which are often involved in actin polymerization.
Function
WH1 domains are important for all cellular processes i ...
and a domain
A domain is a geographic area controlled by a single person or organization. Domain may also refer to:
Law and human geography
* Demesne, in English common law and other Medieval European contexts, lands directly managed by their holder rather ...
that recognises proline
Proline (symbol Pro or P) is an organic acid classed as a proteinogenic amino acid (used in the biosynthesis of proteins), although it does not contain the amino group but is rather a secondary amine. The secondary amine nitrogen is in the p ...
rich sequences (PRS) on other proteins. The EVH1 domain interacts directly with the earlier mentioned NRD of Dcp2, and is currently thought to directly help with the decapping of mRNA, though how it does so is unclear. The domain that recognises PRS is made of mostly hydrophobic
In chemistry, hydrophobicity is the chemical property of a molecule (called a hydrophobe) that is seemingly repelled from a mass of water. In contrast, hydrophiles are attracted to water.
Hydrophobic molecules tend to be nonpolar and, thu ...
amino acids, and is found within the cleft of the 'V' of the Dcp1 structure. It is used to bind to other proteins in the decapping complex to Dcp1.
PNRC2
PNRC2
Proline-rich nuclear receptor coactivator 2 is a protein that in humans is encoded by the ''PNRC2'' gene.
References
Further reading
*
*
*
*
*
*
*
*
External links
*
*
*
Gene expression
Transcription coregulators
{{gene-1-stub ...
attaches to and enhances the effect of Dcp1 to encourage decapping, and also recruits Upf1
Regulator of nonsense transcripts 1 (or Up-frameshift suppressor 1 homolog) is a protein that in humans is encoded by the ''UPF1'' gene.
Function
This gene encodes a protein that is part of a post-splicing multiprotein complex, the exon junct ...
to the decapping complex. It possesses a proline rich sequence that is hydrophobic and sticks strongly to the equally hydrophobic cleft in Dcp1, and so Dcp1 binds PNRC2
Proline-rich nuclear receptor coactivator 2 is a protein that in humans is encoded by the ''PNRC2'' gene.
References
Further reading
*
*
*
*
*
*
*
*
External links
*
*
*
Gene expression
Transcription coregulators
{{gene-1-stub ...
's proline-rich region, which then enhances the function of Dcp2 even more. Current research suggests PNRC2 helps associate Dcp2 and Dcp1 together, making the Dcp2-Dcp1 holoenzyme
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different mol ...
more stable and therefore increasing the effectiveness of Dcp2, but the exact details about how it does so are vague. The recruitment of Upf1
Regulator of nonsense transcripts 1 (or Up-frameshift suppressor 1 homolog) is a protein that in humans is encoded by the ''UPF1'' gene.
Function
This gene encodes a protein that is part of a post-splicing multiprotein complex, the exon junct ...
allows the decapping complex to participate in nonsense-mediated mRNA decay
Nonsense-mediated mRNA decay (NMD) is a surveillance pathway that exists in all eukaryotes. Its main function is to reduce errors in gene expression by eliminating mRNA transcripts that contain premature stop codons. Translation of these aberran ...
, which makes PNRC2 a way for Dcp2 to connect with the regulatory pathway in charge of destroying incorrectly transcripted mRNA.
Upf1-3
Upf1
Regulator of nonsense transcripts 1 (or Up-frameshift suppressor 1 homolog) is a protein that in humans is encoded by the ''UPF1'' gene.
Function
This gene encodes a protein that is part of a post-splicing multiprotein complex, the exon junct ...
, Upf2
Regulator of nonsense transcripts 2 is a protein that in humans is encoded by the ''UPF2'' gene.
Function
This gene encodes a protein that is part of a post-splicing multiprotein complex, the exon junction complex, involved in both mRNA nucle ...
, and Upf3 are proteins involved in the regulatory pathway of nonsense-mediated mRNA decay
Nonsense-mediated mRNA decay (NMD) is a surveillance pathway that exists in all eukaryotes. Its main function is to reduce errors in gene expression by eliminating mRNA transcripts that contain premature stop codons. Translation of these aberran ...
, and not the actual decapping of mRNA. Only Upf1 attaches directly to the decapping complex, whereas Upf2 and Upf3 attach to mRNA, then attach to Upf1 to facilitate the destruction of incorrect mRNA. These are activators of the complex, in that they can direct the complex at incorrectly formed mRNA, but do not actually help decap the mRNA.
DDX6
DDX6, an ortholog of Dhh1, also enhances the effectiveness of the Dcp2-Dcp1 holoenzyme while it hydrolyzes the 5' cap. It is proposed that, since it is ahelicase
Helicases are a class of enzymes that are vital to all organisms. Their main function is to unpack an organism's genetic material. Helicases are motor proteins that move directionally along a nucleic double helix, separating the two hybridized ...
, it is involved in reconfiguring the 5' end
Directionality, in molecular biology and biochemistry, is the end-to-end chemical orientation of a single strand of nucleic acid. In a single strand of DNA or RNA, the chemical convention of naming carbon atoms in the nucleotide pentose-sugar-r ...
of the mRNA to give Dcp2 easier access to the 5' cap, and that it stimulates Dcp1 so that it interacts better with Dcp2 when attached to the rest of the decapping complex.
Edc3
Edc3 further activates the Dcp2-Dcp1 holoenzyme and allows it to quickly decap mRNA. It possesses an LSm domain at itsN-terminus
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the amin ...
, which interacts with specific amino acid motifs called HLM fragments which are found on the C terminus
The C-terminus (also known as the carboxyl-terminus, carboxy-terminus, C-terminal tail, carboxy tail, C-terminal end, or COOH-terminus) is the end of an amino acid chain (protein or polypeptide), terminated by a free carboxyl group (-COOH). When t ...
of Dcp1 and allows for Edc3 to bind to it. Another important part of this protein is the FDF linker, which is a long and unstructured stretch of amino acids that binds with DDX6 and stops it from binding directly with the mRNA, allowing it to interact with the proteins in the decapping complex instead. The final domain of note is a Yjef-N C-terminus domain which dimerizes
In chemistry, dimerization is the process of joining two identical or similar molecular entities by bonds. The resulting bonds can be either strong or weak. Many symmetrical chemical species are described as dimers, even when the monomer is u ...
with mRNA and helps create P-bodies
In cellular biology, P-bodies, or processing bodies, are distinct foci formed by phase separation within the cytoplasm of a eukaryotic cell consisting of many enzymes involved in mRNA turnover. P-bodies are highly conserved structures and have ...
around the location of the decapping complex.
P-bodies
In cellular biology, P-bodies, or processing bodies, are distinct foci formed by phase separation within the cytoplasm of a eukaryotic cell consisting of many enzymes involved in mRNA turnover. P-bodies are highly conserved structures and have ...
are biomolecular condensates rich in decapped or repressed mRNA mixed together with mRNA degradation factors, such as the decapping complex and the nonsense-mediated mRNA decay machinery, so they are important for the eventual destruction of the mRNA altered by Dcp2. As Edc3 creates P-bodies around the decapping complex, it becomes easier for Dcp2 to access the mRNA 5' caps to hydrolyze, increasing the effectiveness of the entire complex.
Pat1
Pat1 is another protein that increases the efficiency of the decapping complex. It has three main domains. One is necessary for decapping mRNA, and directly helps the Dcp2-Dcp1 holoenzyme do so. The other two make it easier for the protein to decap mRNA, but are not directly involved in the hydrolysis of the phosphate bond. Pat1 enhances binding of the 3′ complex to RNA and activates the 5′ decapping complex by multiple mechanisms including promoting phase separation. Pat1 has many interactions with the various proteins in the decapping complex, and is known as the 'scaffolding protein' because it brings everything together when it is time to decap something. The N-terminus domain interacts with DXX6 and brings it close so it can activate Dcp1, another portion helps create P-bodies along with Edc3, and the C-terminus domains attach Dcp1–Dcp2, the Lsm1–7 complex andXrn1
5′-3′ exoribonuclease 1 (Xrn1) is a protein that in humans is encoded by the XRN1 gene. Xrn1 hydrolyses RNA in the 5′ to 3′ direction.
Function
This gene encodes a member of the 5′-3′ exonuclease family. The encoded protein may be ...
to the complex.

Xrn1
Xrn1
5′-3′ exoribonuclease 1 (Xrn1) is a protein that in humans is encoded by the XRN1 gene. Xrn1 hydrolyses RNA in the 5′ to 3′ direction.
Function
This gene encodes a member of the 5′-3′ exonuclease family. The encoded protein may be ...
is a 5' to 3' exonuclease that degrades the just-decapped mRNA. It targets the 5' monophosphate end of mRNA, which is what is left over when Dcp2 has hydrolyzed the cap off and taken away the 7-methylguanosine
7-Methylguanosine (m7G) is a modified purine nucleoside. It is a methylated version of guanosine and when found in human urine, it may be a biomarker of some types of cancer. In the RNAs, 7-methylguanosine have been used to study and examine t ...
cap, along with two of the three phosphates that attach the cap to the mRNA. The current theory is that the structure of Xrn1 does not allow a capped mRNA to interact with it because the Xrn1 is structured in such a way that there is steric hindrance
Steric effects arise from the spatial arrangement of atoms. When atoms come close together there is generally a rise in the energy of the molecule. Steric effects are nonbonding interactions that influence the shape ( conformation) and reactivi ...
that physically blocks the protein from interacting with any mRNA that Dcp2 has not already decapped.
References
{{reflist RNA