Circular RNA (or circRNA) is a type of single-stranded
RNA which, unlike linear RNA, forms a
covalently
A covalent bond is a chemical bond that involves the sharing of electrons to form electron pairs between atoms. These electron pairs are known as shared pairs or bonding pairs. The stable balance of attractive and repulsive forces between atoms ...
closed continuous loop. In circular RNA, the
3' and 5' ends normally present in an RNA molecule have been joined together. This feature confers numerous properties to circular RNA, many of which have only recently been identified.
Many types of circular RNA arise from otherwise protein-coding genes. Some circular RNA has been shown to code for proteins. Some types of circular RNA have also recently shown potential as gene regulators. The biological function of most circular RNA is unclear.
Because circular RNA does not have 5' or 3' ends, it is resistant to
exonuclease
Exonucleases are enzymes that work by cleaving nucleotides one at a time from the end (exo) of a polynucleotide chain. A hydrolyzing reaction that breaks phosphodiester bonds at either the 3′ or the 5′ end occurs. Its close relative is t ...
-mediated degradation and is presumably more stable than most linear RNA in cells.
Circular RNA has been linked to some diseases such as cancer.
RNA splicing
In contrast to genes in
bacteria,
eukaryotic genes are split by non-coding sequences called
introns. In eukaryotes, as a gene is transcribed from DNA into a
messenger RNA (mRNA) transcript, intervening introns are removed, leaving only
exons in the mature mRNA, which can subsequently be translated to produce the protein product.
The
spliceosome,
a protein-RNA complex located in the nucleus, catalyzes splicing in the following manner:
# The
spliceosome recognizes an
intron, which is flanked by specific sequences at its 5' and 3' ends, known as a donor splice site (or 5' splice site) and an acceptor splice site (or 3' splice site), respectively.
# The 5' splice site sequence is then subjected to a
nucleophilic
In chemistry, a nucleophile is a chemical species that forms bonds by donating an electron pair. All molecules and ions with a free pair of electrons or at least one pi bond can act as nucleophiles. Because nucleophiles donate electrons, they are ...
attack by a downstream sequence called the branch point, resulting in a circular structure called a lariat.
# The free 5' exon then attacks the 3' splice site, joining the two exons and releasing a structure known as an ''intron lariat''. The intron lariat is subsequently de-branched and quickly degraded.

Alternative splicing
Alternative splicing
Alternative splicing, or alternative RNA splicing, or differential splicing, is an alternative splicing process during gene expression that allows a single gene to code for multiple proteins. In this process, particular exons of a gene may be i ...
is a phenomenon through which one RNA transcript can yield different protein products based on which segments are considered "introns" and "exons" during a splicing event.
Although not specific to humans, it is a partial explanation for the fact that humans and other much simpler species (such as nematodes) have similar numbers of genes (in the range of 20 - 25 thousand). One of the most striking examples of alternative splicing is in the ''Drosophila''
DSCAM gene, which can give rise to approximately 30 thousand distinct alternatively spliced isoforms.
Non-canonical splicing
Exon scrambling
Exon scrambling, also called exon shuffling, describes an event in which exons are spliced in a "non-canonical" (atypical) order. There are three ways in which exon scrambling can occur:
#
Tandem exon duplication Tandem exon duplication is defined as duplication of exons within the same gene to give rise to the subsequent exon. A complete exon analysis of all genes in ''Homo sapiens'', ''Drosophila melanogaster'', and ''Caenorhabditis elegans'' has shown 12, ...
in the genome, which often occurs in cancers.
#
Trans-splicing
''Trans''-splicing is a special form of RNA processing where exons from two different primary RNA transcripts are joined end to end and ligated. It is usually found in eukaryotes and mediated by the spliceosome, although some bacteria and archaea ...
, in which two RNA transcripts fuse, resulting in a linear transcript containing exons that, for example, may be derived from genes encoded on two different chromosomes. Trans-splicing is very common in ''C. elegans''
# A splice donor site being joined to a splice acceptor site further upstream in the primary transcript, yielding a circular transcript.
The notion that circularized transcripts are byproducts from imperfect splicing is supported by the low abundance and the lack of sequence conservation of most circRNAs,
but has been challenged.
[
]
Alu element
An Alu element is a short stretch of DNA originally characterized by the action of the ''Arthrobacter luteus (Alu)'' restriction endonuclease. ''Alu'' elements are the most abundant transposable elements, containing over one million copies dis ...
s impact circRNA splicing
Repetitive Alu sequences represent approximately 10% of the human genome. The presence of Alu elements in flanking introns of protein-coding genes adjacent to the first and last exons that form circRNAs, influence the formation of circRNAs.[ It is important that the flanking intronic ]Alu element
An Alu element is a short stretch of DNA originally characterized by the action of the ''Arthrobacter luteus (Alu)'' restriction endonuclease. ''Alu'' elements are the most abundant transposable elements, containing over one million copies dis ...
s are complementary, as this enables RNA pairing, which in turn facilitates circRNA synthesis.
Impact of RNA editing on circRNA formation
RNAs can undergo base modification by RNA editing after transcription. RNA editing occurs mainly in Alu element
An Alu element is a short stretch of DNA originally characterized by the action of the ''Arthrobacter luteus (Alu)'' restriction endonuclease. ''Alu'' elements are the most abundant transposable elements, containing over one million copies dis ...
s of protein-coding genes.Alu element
An Alu element is a short stretch of DNA originally characterized by the action of the ''Arthrobacter luteus (Alu)'' restriction endonuclease. ''Alu'' elements are the most abundant transposable elements, containing over one million copies dis ...
s flanking the back-splice site (BSS) can reduces the formation of circRNAs in the human heart.Alu element
An Alu element is a short stretch of DNA originally characterized by the action of the ''Arthrobacter luteus (Alu)'' restriction endonuclease. ''Alu'' elements are the most abundant transposable elements, containing over one million copies dis ...
s flanking the back-splice site.
Characteristics of circular RNA
Early discoveries of circRNAs
Early discoveries of circular RNAs led to the belief that they lacked significance due to their rarity. These early discoveries included the analysis of genes like the DCC and Sry genes, and the recent discovery of the human non-coding RNA ANRIL, all of which expressed circular isoforms. CircRNA producing genes like the human ETS-1 gene, the human and rat cytochrome P450
Cytochromes P450 (CYPs) are a superfamily of enzymes containing heme as a cofactor that functions as monooxygenases. In mammals, these proteins oxidize steroids, fatty acids, and xenobiotics, and are important for the clearance of various comp ...
genes, the rat androgen binding protein gene (Shbg
Sex hormone-binding globulin (SHBG) or sex steroid-binding globulin (SSBG) is a glycoprotein that binds to androgens and estrogens. When produced by the Sertoli cells in the seminiferous tubules of the testis, it has also been called androgen- ...
), and the human dystrophin gene were also discovered.
Genome-wide identification of circRNAs
Scrambled isoforms and circRNAs
In 2012, in an effort to initially identify cancer-specific exon scrambling events, scrambled exons were discovered in large numbers in both normal and cancer cells. It was found that scrambled exon isoforms comprised about 10% of the total transcript isoforms in leukocytes, with 2,748 scrambled isoforms in HeLa and H9 embryonic stem cells being identified. Additionally, about 1 in 50 expressed genes produced scrambled transcript isoforms at least 10% of the time. Tests used to recognize circularity included treating samples with RNase R RNase R, or Ribonuclease R, is a 3'-->5' exoribonuclease, which belongs to the RNase II superfamily, a group of enzymes that hydrolyze RNA in the 3' - 5' direction. RNase R has been shown to be involved in selective mRNA degradation, particularly ...
, an enzyme that degrades linear but not circular RNAs, and testing for the presence of poly-A tails, which are not present in circular molecules. Overall, 98% of scrambled isoforms were found to represent circRNAs, circRNAs were found to be located in the cytoplasm, and circRNAs were found to be abundant.
Discovery of a higher abundance of circRNAs
In 2013, a higher abundance of circRNAs was discovered. Human fibroblast RNA was treated with RNase R to enrich for circular RNAs, followed by the categorization of circular transcripts based on their abundance (low, medium, high).[ Approximately 1 in 8 expressed genes were found to produce detectable levels of circRNAs, including those of low abundance, which was significantly higher than previously suspected, and was attributed to greater sequencing depth.]
CircRNAs tissue specificity and antagonist activity
At the same time, a computational method to detect circRNAs was developed, leading to de novo detection of circRNAs in humans, mice, and ''C. elegans'', and extensively validating them. The expression of circRNAs was often found to be tissue/developmental stage specific. Additionally, circRNAs were found to have the ability to act as antagonists of miRNAs, microRNAs which interfere with translation of mRNAs, as exemplified by the circRNA CDR1as, which has miRNA binding sites (as seen below).
CircRNAs and ENCODE Ribozero RNA-seq data
In 2014, human circRNAs were identified and quantified from ENCODE Ribozero RNA-seq data. Most circRNAs were found to be minor splice isoforms and to be expressed in only a few cell types, with 7,112 human circRNAs having circular fractions (the fraction of similarity an isoform has to transcripts the same locus) of at least 10%. CircRNAs were also found to be no more conserved than their linear controls and, according to ribosome profiling, are not translated.<
CircRNAs and CIRCexplorer
In the same year, CIRCexplorer, a tool used to identify thousands of circRNAs in humans without RNase R RNA-seq data, was developed. The vast majority of identified highly expressed exonic circular RNAs were found to be processed from exons located in the middle of RefSeq
The Reference Sequence (RefSeq) database is an open access, annotated and curated collection of publicly available nucleotide sequences ( DNA, RNA) and their protein products. RefSeq was first introduced in 2000. This database is built by National ...
genes, suggesting that the circular RNA formation is generally coupled to RNA splicing. It was determined that most circular RNAs contain multiple, most commonly, two to three, exons. Exons from circRNAs with only one circularized exon were found to be much longer than those from circRNAs with multiple circularized exons, indicating that processing may prefer a certain length to maximize exon(s) circularization. The introns of circularized exons generally contain high Alu densities that can form inverted repeated Alu pairs (IRAlus). IRAlus, either convergent or divergent, are juxtaposed across flanking introns of circRNAs in a parallel way with similar distances to adjacent exons. IRAlus, and other non-repetitive, but complementary, sequences were also found to promote circular RNA formation. On the other hand, exon circularization efficiency was determined to be affected by the competition of RNA pairing, such that alternative RNA pairing, and its competition, leads to alternative circularization. Finally, both exon circularization and its regulation were found to be evolutionarily dynamic.[
]
Genome-wide calling of circRNA in Alzheimer disease cases
The Cruchaga lab performed the first large scale analyses of circRNA in Alzheimer disease (AD) and demonstrated the role of circRNAs in health and disease. A total of 148 circRNAs were found to be significantly associated in multiple datasets with Alzheimer's disease
Alzheimer's disease (AD) is a neurodegenerative disease that usually starts slowly and progressively worsens. It is the cause of 60–70% of cases of dementia. The most common early symptom is difficulty in remembering recent events. As ...
status and clinical dementia rating
The Clinical Dementia Rating or CDR is a numeric scale used to quantify the severity of symptoms of dementia (i.e. its 'stage').
Scale
Using a structured-interview protocol developed by Charles Hughes, Leonard Berg, John C. Morris and other colle ...
(CDR) at death after false discovery rate (FDR) correction. The expression of circRNAs was independent of the lineal form and that circRNA expression was also corrected by cell proportion. CircRNAs were also found to be co-expressed with known causal Alzheimer genes, such as APP and PSEN1
Presenilin-1 (PS-1) is a presenilin protein that in humans is encoded by the ''PSEN1'' gene. Presenilin-1 is one of the four core proteins in the gamma secretase complex, which is considered to play an important role in generation of amyloid beta ...
, indicating that some circRNAs are also part of the causal pathway. Altogether, circRNA brain expression was found to explain more about Alzheimer’s clinical manifestations than the number of APOε4 alleles, suggesting that circRNAs could be used as a potential biomarker for Alzheimer’s.
Classes of CircRNA
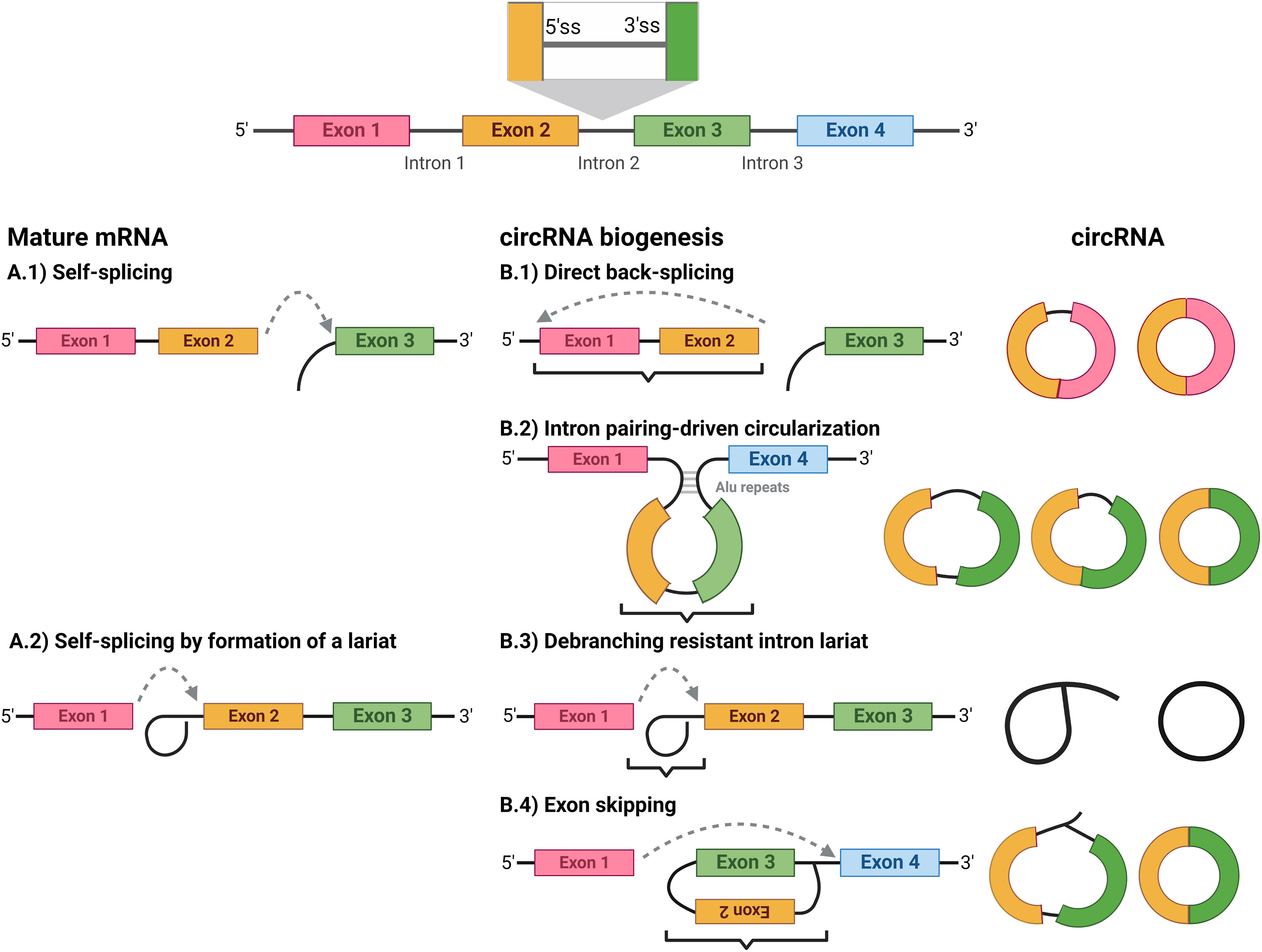
 Circular RNAs can be separated into five classes:
Circular RNAs can be separated into five classes:
Length of circRNAs
A recent study of human circRNAs revealed that these molecules are usually composed of 1–5 exons.
Location of circRNAs in the cell
In the cell, circRNAs are predominantly found in the cytoplasm
In cell biology, the cytoplasm is all of the material within a eukaryotic cell, enclosed by the cell membrane, except for the cell nucleus. The material inside the nucleus and contained within the nuclear membrane is termed the nucleoplasm. Th ...
, where the number of circular RNA transcripts derived from a gene can be up to ten times greater than the number of associated linear RNAs generated from that locus
Locus (plural loci) is Latin for "place". It may refer to:
Entertainment
* Locus (comics), a Marvel Comics mutant villainess, a member of the Mutant Liberation Front
* ''Locus'' (magazine), science fiction and fantasy magazine
** ''Locus Award' ...
. It is unclear how circular RNAs exit the nucleus
Nucleus ( : nuclei) is a Latin word for the seed inside a fruit. It most often refers to:
* Atomic nucleus, the very dense central region of an atom
*Cell nucleus, a central organelle of a eukaryotic cell, containing most of the cell's DNA
Nucl ...
through a relatively small nuclear pore
A nuclear pore is a part of a large complex of proteins, known as a nuclear pore complex that spans the nuclear envelope, which is the double membrane surrounding the eukaryotic cell nucleus. There are approximately 1,000 nuclear pore complexes ...
. Because the nuclear envelope breaks down during mitosis, one hypothesis is that the molecules exit the nucleus during this phase of the cell cycle
The cell cycle, or cell-division cycle, is the series of events that take place in a cell that cause it to divide into two daughter cells. These events include the duplication of its DNA (DNA replication) and some of its organelles, and subs ...
.
CircRNAs are stable compared to linear RNAs
CircRNAs lack a polyadenylated tail and, therefore, are predicted to be less prone to degradation by exonucleases. In 2015, Enuka ''et al.'' measured the half-lives of 60 circRNAs and their linear counterparts expressed from the same host gene and revealed that the median half-life of circRNAs of mammary cells (18.8 to 23.7 hours) is at least 2.5 times longer than the median half-life of their linear counterparts (4.0 to 7.4 hours).
Plausible functions of circular RNA
Evolutionary conservation of circularization mechanisms and signals
CircRNAs have been identified in various species across the domains of life. In 2011, Danan ''et al.'' sequenced RNA from Archaea. After digesting total RNA with RNase R, they were able to identify circular species, indicating that circRNAs are not specific to eukaryotes. However, these archaeal circular species are probably not made via splicing, suggesting that other mechanisms to generate circular RNA likely exist.
CircRNAs were found to be largely conserved between human and sheep. By analyzing total RNA sequencing data from sheep's parietal lobe cortex and peripheral blood mononuclear cells it was shown that 63% of the detected circRNAs are homologous to known human circRNAs.
 In a closer evolutionary connection, a comparison of RNA from mouse testes vs. RNA from a human cell found 69 orthologous circRNAs. For example, both humans and mice encode the
In a closer evolutionary connection, a comparison of RNA from mouse testes vs. RNA from a human cell found 69 orthologous circRNAs. For example, both humans and mice encode the HIPK2
Homeodomain-interacting protein kinase 2 is an enzyme that in humans is encoded by the ''HIPK2'' gene. HIPK2 can be categorized as a Serine/Threonine Protein kinase, specifically one that interacts with homeodomain transcription factors. It belon ...
and HIPK3 genes, two paralogous
Sequence homology is the biological homology between DNA, RNA, or protein sequences, defined in terms of shared ancestry in the evolutionary history of life. Two segments of DNA can have shared ancestry because of three phenomena: either a spe ...
kinases which produce a large amount of circRNA from one particular exon in both species.
CDR1as/CiRS-7 as a miR-7 sponge
microRNA
MicroRNA (miRNA) are small, single-stranded, non-coding RNA molecules containing 21 to 23 nucleotides. Found in plants, animals and some viruses, miRNAs are involved in RNA silencing and post-transcriptional regulation of gene expression. miRN ...
s (miRNAs) are small (~21nt) non-coding RNAs that repress translation of messenger RNAs involved in a large, diverse set of biological processes. They directly base-pair to target messenger RNAs (mRNAs), and can trigger cleavage of the mRNA depending on the degree of complementarity.
MicroRNAs are grouped in "seed families." Family members share nucleotides 2–7, known as the seed region.Argonaute
The Argonaute protein family, first discovered for its evolutionarily conserved stem cell function, plays a central role in RNA silencing processes as essential components of the RNA-induced silencing complex (RISC). RISC is responsible for the ...
proteins are the "effector proteins" which help miRNAs carry out their job, while microRNA sponges are RNAs that "sponge up" miRNAs of a particular family, thereby serving as competitive inhibitors that suppress the ability of the miRNA to bind its mRNA targets, thanks to the presence of multiple binding sites that recognize a specific seed region.antisense
In molecular biology and genetics, the sense of a nucleic acid molecule, particularly of a strand of DNA or RNA, refers to the nature of the roles of the strand and its complement in specifying a sequence of amino acids. Depending on the context, ...
to the human CDR1 (gene) locus (hence the name CDR1as),zebrafish
The zebrafish (''Danio rerio'') is a freshwater fish belonging to the minnow family (Cyprinidae) of the order Cypriniformes. Native to South Asia, it is a popular aquarium fish, frequently sold under the trade name zebra danio (and thus often ...
, which do not have the CDR1 locus in their genome, provides evidence for CiRS-7's sponge activity. During development, miR-7 is strongly expressed in the zebrafish brain. To silence miR-7 expression in zebrafish, Memczak and colleagues took advantage of a tool called morpholino
A Morpholino, also known as a Morpholino oligomer and as a phosphorodiamidate Morpholino oligomer (PMO), is a type of oligomer molecule (colloquially, an oligo) used in molecular biology to modify gene expression. Its molecular structure contains ...
, which can base pair and sequester target molecules. Morpholino treatment had the same severe effect on midbrain development as ectopically expressing CiRS-7 in zebrafish brains using injected plasmids. This indicates a significant interaction between CiRS-7 and miR-7 in vivo.inverted repeat An inverted repeat (or IR) is a single stranded sequence of nucleotides followed downstream by its reverse complement. The intervening sequence of nucleotides between the initial sequence and the reverse complement can be any length including zero. ...
s (IRs) over 15.5 kilobases (kb) in length. When one or both of the IRs are deleted, circularization does not occur. It was this finding that introduced the idea of inverted repeats enabling circularization.
Because circular RNA sponges are characterized by high expression levels, stability, and a large number of miRNA binding sites, they are likely to be more effective sponges than those that are linear.
Other possible functions for circRNAs
Though recent attention has been focused on circRNA's "sponge" functions, scientists are considering several other functional possibilities as well. For example, some areas of the mouse adult hippocampus show expression of CiRS-7 but not miR-7, suggesting that CiRS-7 may have roles that are independent of interacting with the miRNA.RNA-binding protein
RNA-binding proteins (often abbreviated as RBPs) are proteins that bind to the double or single stranded RNA in cells and participate in forming ribonucleoprotein complexes.
RBPs contain various structural motifs, such as RNA recognition motif ...
s (RBPs) and RNAs besides miRNAs to form RNA-protein complexes.
Circular intronic long non-coding RNAs (ciRNAs)
Usually, intronic lariats (see above) are debranched and rapidly degraded. However, a debranching failure can lead to the formation of circular intronic long non-coding RNAs, also known as ciRNAs.nucleus
Nucleus ( : nuclei) is a Latin word for the seed inside a fruit. It most often refers to:
* Atomic nucleus, the very dense central region of an atom
*Cell nucleus, a central organelle of a eukaryotic cell, containing most of the cell's DNA
Nucl ...
rather than the cytoplasm
In cell biology, the cytoplasm is all of the material within a eukaryotic cell, enclosed by the cell membrane, except for the cell nucleus. The material inside the nucleus and contained within the nuclear membrane is termed the nucleoplasm. Th ...
. In addition, these molecules contain few (if any) miRNA binding sites. Instead of acting as sponges, ciRNAs seem to function in regulating the expression of their parent genes. For example, a relatively abundant ciRNA called ci-ankrd52 positively regulates Pol II transcription. Many ciRNAs remain at their "sites of synthesis" in the nucleus. However, ciRNA may have roles other than simply regulating their parent genes, as ciRNAs do localize to additional sites in the nucleus other than their "sites of synthesis".
Circular RNA and disease
As with most topics in molecular biology, it is important to consider how circular RNA can be used as a tool to help mankind. Given its abundance, evolutionary conservation, and potential regulatory role, it is worthwhile to look into how circular RNA can be used to study pathogenesis and devise therapeutic interventions. For example:
* Circular ANRIL (cANRIL) is the circular form of ANRIL, a long non-coding RNA (ncRNA). Expression of cANRIL is correlated with risk for atherosclerosis
Atherosclerosis is a pattern of the disease arteriosclerosis in which the wall of the artery develops abnormalities, called lesions. These lesions may lead to narrowing due to the buildup of atheromatous plaque. At onset there are usually no ...
, a disease in which the arteries become hard. It has been proposed that cANRIL can modify INK4/ARF expression, which, in turn, increases risk for atherosclerosis. Further study of cANRIL expression could potentially be used to prevent or treat atherosclerosis.
* miR-7 plays an important regulatory role in several cancers and in Parkinson's disease, which is a brain illness of unknown origin.transgene
A transgene is a gene that has been transferred naturally, or by any of a number of genetic engineering techniques, from one organism to another. The introduction of a transgene, in a process known as transgenesis, has the potential to change t ...
, which is a synthetic gene that is transferred between organisms. It is also important to consider how transgenes can be expressed only in specific tissues, or expressed only when induced.
Circular RNAs play a role in Alzheimer disease pathogenesis
Dube et al.,[ demonstrated for the first time that brain circular RNAs (circRNA) are part of the pathogenic events that lead to ]Alzheimer’s disease
Alzheimer's disease (AD) is a neurodegenerative disease that usually starts slowly and progressively worsens. It is the cause of 60–70% of cases of dementia. The most common early symptom is difficulty in remembering recent events. As t ...
, hypothesizing that specific circRNA would be differentially expressed in AD cases compared to controls and that those effects could be detected early in the disease. They optimized and validated a novel analyses pipeline for circular RNAs (circRNA). They performed a three-stage study design, using the Knight ADRC brain RNA-seq data as discovery (stage 1), using the data from Mount Sinai as replication (stage 2) and a meta-analysis (stage 3) to identify the most significant circRNA differentially expressed in Alzheimer disease. Using his pipeline, they found 3,547 circRNA that passed stringent QC in the Knight ADRC cohort that includes RNA-seq from 13 controls and 83 Alzheimer cases, and 3,924 circRNA passed stringent QC in the MSBB dataset. A meta-analysis of the discovery and replication results revealed a total of 148 circRNAs that were significantly correlated with CDR after FDR correction. In addition, 33 circRNA passed the stringent gene-based, Bonferroni multiple test correction of 5×10-6, including circHOMER1 (P =2.21×10−18) and circCDR1-AS (P = 2.83 × 10−8), among others. They also performed additional analyses to demonstrate that the expression of circRNA were independent of the lineal form as well as the cell proportion that can confound the brain RNA-seq analyses in Alzheimer disease studies. They performed co-expression analyses of all the circRNA together with the lineal forms and found that circRNA, including those that were differentially expressed in Alzheimer disease compared to controls co-expressed with known causal Alzheimer genes, such as APP and PSEN1, indicating that some circRNA are also part of the causal pathway. They also demonstrated that cirRNA brain expression explained more about Alzheimer clinical manifestations that the number of APOε4 alleles, suggesting that could be used as a potential biomarker for Alzheimer disease. This is an important study for the field, as it is the first time that circRNA are quantified and validated (by real-time PCR) in human brain samples at genome-wide scale and in large and well-characterized cohorts. It also demonstrates that these RNA forms are likely to be implicated on complex traits including Alzheimer disease will help to understand the biological events that leads to disease.
Viroids as circular RNAs
Viroids are mostly plant pathogens, which consist of short stretches (a few hundred nucleobases) of highly complementary, circular, single-stranded, and non-coding RNAs without a protein coat. Compared with other infectious plant pathogens, viroids are extremely small in size, ranging from 246 to 467 nucleobases; they thus consist of fewer than 10,000 atoms. In comparison, the genome of the smallest known viruses capable of causing an infection by themselves are around 2,000 nucleobases long.
See also
* Circular RNA (circRNA) databases and resources
References
{{reflist, 30em
RNA
 Circular RNA (or circRNA) is a type of single-stranded RNA which, unlike linear RNA, forms a
Circular RNA (or circRNA) is a type of single-stranded RNA which, unlike linear RNA, forms a  Circular RNAs can be separated into five classes:
Circular RNAs can be separated into five classes: