Cas9 on:
[Wikipedia]
[Google]
[Amazon]
Cas9 (CRISPR associated protein 9, formerly called Cas5, Csn1, or Csx12) is a 160
 CRISPR-Cas systems are divided into three major types (type I, type II, and type III) and twelve subtypes, which are based on their genetic content and structural differences. However, the core defining features of all CRISPR-Cas systems are the cas genes and their proteins: cas1 and cas2 are universal across types and subtypes, while
CRISPR-Cas systems are divided into three major types (type I, type II, and type III) and twelve subtypes, which are based on their genetic content and structural differences. However, the core defining features of all CRISPR-Cas systems are the cas genes and their proteins: cas1 and cas2 are universal across types and subtypes, while
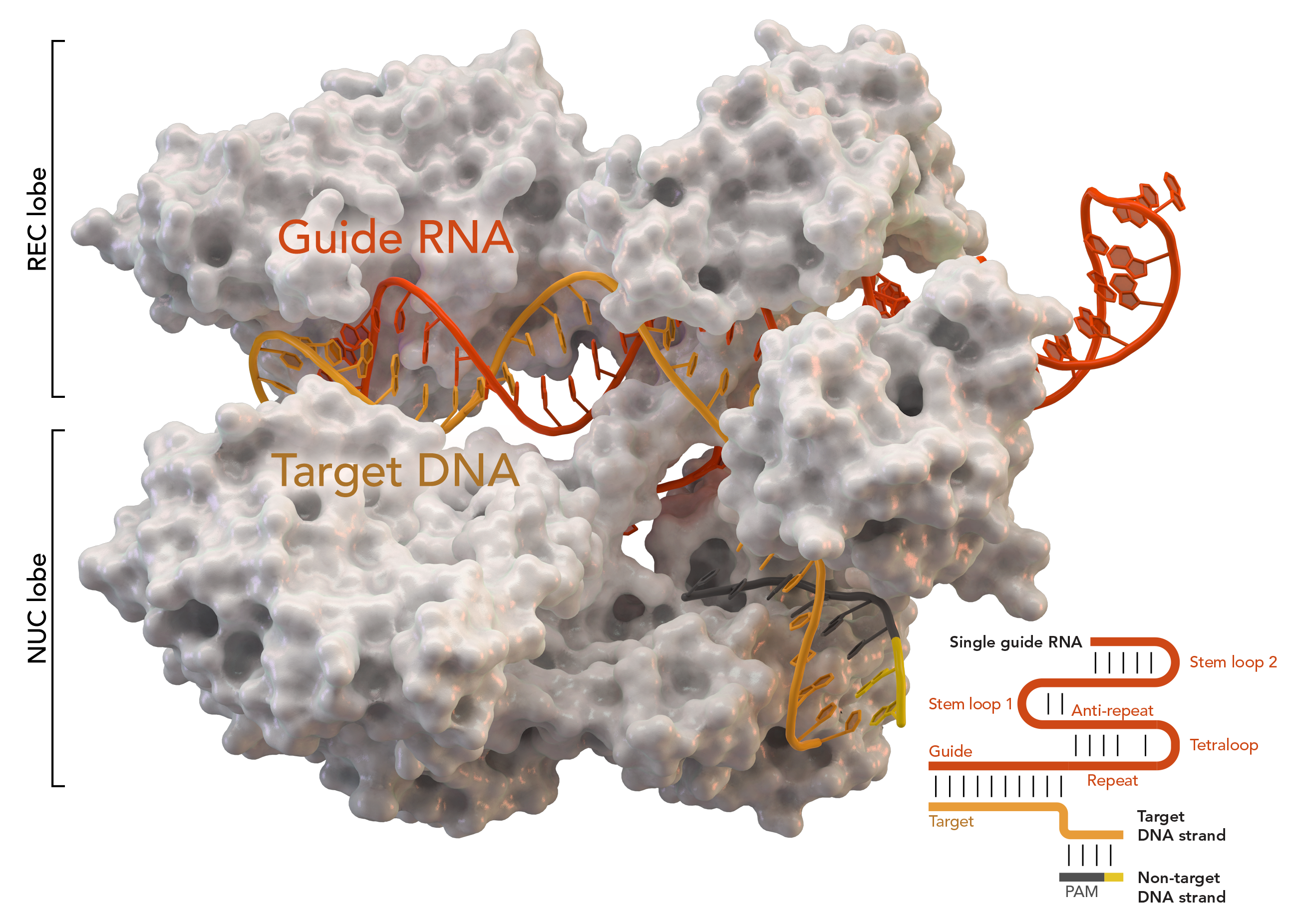 Cas9 features a bi-lobed architecture with the guide RNA nestled between the alpha-helical lobe (blue) and the nuclease lobe (cyan, orange, and gray). These two lobes are connected through a single bridge helix. There are two nuclease domains located in the multi-domain nuclease lobe, the RuvC (gray) which cleaves the non-target DNA strand, and the HNH nuclease domain (cyan) that cleaves the target strand of DNA. The RuvC domain is encoded by sequentially disparate sites that interact in the tertiary structure to form the RuvC cleavage domain (See right figure).
Cas9 features a bi-lobed architecture with the guide RNA nestled between the alpha-helical lobe (blue) and the nuclease lobe (cyan, orange, and gray). These two lobes are connected through a single bridge helix. There are two nuclease domains located in the multi-domain nuclease lobe, the RuvC (gray) which cleaves the non-target DNA strand, and the HNH nuclease domain (cyan) that cleaves the target strand of DNA. The RuvC domain is encoded by sequentially disparate sites that interact in the tertiary structure to form the RuvC cleavage domain (See right figure).
 A key feature of the target DNA is that it must contain a protospacer adjacent motif (PAM) consisting of the three-nucleotide sequence- NGG. This PAM is recognized by the PAM-interacting domain (PI domain, orange) located near the C-terminal end of Cas9. Cas9 undergoes distinct conformational changes between the apo, guide RNA bound, and guide RNA:DNA bound states.
Cas9 recognizes the
A key feature of the target DNA is that it must contain a protospacer adjacent motif (PAM) consisting of the three-nucleotide sequence- NGG. This PAM is recognized by the PAM-interacting domain (PI domain, orange) located near the C-terminal end of Cas9. Cas9 undergoes distinct conformational changes between the apo, guide RNA bound, and guide RNA:DNA bound states.
Cas9 recognizes the
 In sgRNA-Cas9 complex, based on the crystal structure, REC1, BH and PI domains have important contacts with backbone or bases in both repeat and spacer region. Several Cas9 mutants including REC1 or REC2 domains deletion and residues mutations in BH have been tested. REC1 and BH related mutants show lower or none activity compared with wild type, which indicate these two domains are crucial for the sgRNA recognition at repeat sequence and stabilization of the whole complex. Although the interactions between spacer sequence and Cas9 as well as PI domain and repeat region need further studies, the co-crystal demonstrates clear interface between Cas9 and sgRNA.
In sgRNA-Cas9 complex, based on the crystal structure, REC1, BH and PI domains have important contacts with backbone or bases in both repeat and spacer region. Several Cas9 mutants including REC1 or REC2 domains deletion and residues mutations in BH have been tested. REC1 and BH related mutants show lower or none activity compared with wild type, which indicate these two domains are crucial for the sgRNA recognition at repeat sequence and stabilization of the whole complex. Although the interactions between spacer sequence and Cas9 as well as PI domain and repeat region need further studies, the co-crystal demonstrates clear interface between Cas9 and sgRNA.
 Previous sequence analysis and biochemical studies have posited that Cas9 contains two nuclease domains: an McrA-like HNH nuclease domain and a RuvC-like nuclease domain. These HNH and RuvC-like nuclease domains are responsible for cleavage of the complementary/target and non-complementary/non-target DNA strands, respectively. Despite low sequence similarity, the sequence similar to RNase H has a RuvC fold (one member of RNase H family) and the HNH region folds as T4 Endo VII (one member of HNH endonuclease family).
Wild-type ''S. pyogenes'' Cas9 requires magnesium (Mg2+)
Previous sequence analysis and biochemical studies have posited that Cas9 contains two nuclease domains: an McrA-like HNH nuclease domain and a RuvC-like nuclease domain. These HNH and RuvC-like nuclease domains are responsible for cleavage of the complementary/target and non-complementary/non-target DNA strands, respectively. Despite low sequence similarity, the sequence similar to RNase H has a RuvC fold (one member of RNase H family) and the HNH region folds as T4 Endo VII (one member of HNH endonuclease family).
Wild-type ''S. pyogenes'' Cas9 requires magnesium (Mg2+)
kilodalton
The dalton or unified atomic mass unit (symbols: Da or u) is a non-SI unit of mass widely used in physics and chemistry. It is defined as of the mass of an unbound neutral atom of carbon-12 in its nuclear and electronic ground state and at ...
protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, respo ...
which plays a vital role in the immunological defense of certain bacteria against DNA virus
A DNA virus is a virus that has a genome made of deoxyribonucleic acid (DNA) that is replicated by a DNA polymerase. They can be divided between those that have two strands of DNA in their genome, called double-stranded DNA (dsDNA) viruses, and ...
es and plasmid
A plasmid is a small, extrachromosomal DNA molecule within a cell that is physically separated from chromosomal DNA and can replicate independently. They are most commonly found as small circular, double-stranded DNA molecules in bacteria; how ...
s, and is heavily utilized in genetic engineering
Genetic engineering, also called genetic modification or genetic manipulation, is the modification and manipulation of an organism's genes using technology. It is a set of technologies used to change the genetic makeup of cells, including t ...
applications. Its main function is to cut DNA and thereby alter a cell's genome. The CRISPR-Cas9 genome editing technique was a significant contributor to the Nobel Prize in Chemistry
)
, image = Nobel Prize.png
, alt = A golden medallion with an embossed image of a bearded man facing left in profile. To the left of the man is the text "ALFR•" then "NOBEL", and on the right, the text (smaller) "NAT•" then "M ...
in 2020 being awarded to Emmanuelle Charpentier
Emmanuelle Marie Charpentier (; born 11 December 1968) is a French professor and researcher in microbiology, genetics, and biochemistry. As of 2015, she has been a director at the Max Planck Institute for Infection Biology in Berlin. In 2018, sh ...
and Jennifer Doudna
Jennifer Anne Doudna (; born February 19, 1964) is an American biochemist who has done pioneering work in CRISPR gene editing, and made other fundamental contributions in biochemistry and genetics. Doudna was one of the first women to share a ...
.
More technically, Cas9 is a dual RNA
Ribonucleic acid (RNA) is a polymeric molecule essential in various biological roles in coding, decoding, regulation and expression of genes. RNA and deoxyribonucleic acid ( DNA) are nucleic acids. Along with lipids, proteins, and carbohydra ...
-guided DNA endonuclease
Endonucleases are enzymes that cleave the phosphodiester bond within a polynucleotide chain. Some, such as deoxyribonuclease I, cut DNA relatively nonspecifically (without regard to sequence), while many, typically called restriction endonucleases ...
enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. A ...
associated with the Clustered Regularly Interspaced Short Palindromic Repeats ( CRISPR) adaptive immune system in ''Streptococcus pyogenes
''Streptococcus pyogenes'' is a species of Gram-positive, aerotolerant bacteria in the genus ''Streptococcus''. These bacteria are extracellular, and made up of non-motile and non-sporing cocci (round cells) that tend to link in chains. They are ...
''. ''S. pyogenes'' utilizes CRISPR to memorize and Cas9 to later interrogate and cleave foreign DNA, such as invading bacteriophage
A bacteriophage (), also known informally as a ''phage'' (), is a duplodnaviria virus that infects and replicates within bacteria and archaea. The term was derived from "bacteria" and the Greek φαγεῖν ('), meaning "to devour". Bacteri ...
DNA or plasmid DNA. Cas9 performs this interrogation by unwinding foreign DNA and checking for sites complementary to the 20 nucleotide spacer region of the guide RNA
A guide RNA (gRNA) is a piece of RNA that functions as a guide for RNA- or DNA-targeting enzymes, with which it forms complexes. Very often these enzymes will delete, insert or otherwise alter the targeted RNA or DNA. They occur naturally, se ...
(gRNA). If the DNA substrate is complementary to the guide RNA, Cas9 cleaves the invading DNA. In this sense, the CRISPR-Cas9 mechanism has a number of parallels with the RNA interference
RNA interference (RNAi) is a biological process in which RNA molecules are involved in sequence-specific suppression of gene expression by double-stranded RNA, through translational or transcriptional repression. Historically, RNAi was known by o ...
(RNAi) mechanism in eukaryotes.
Apart from its original function in bacterial immunity, the Cas9 protein has been heavily utilized as a genome engineering tool to induce site-directed double-strand breaks in DNA. These breaks can lead to gene inactivation or the introduction of heterologous genes through non-homologous end joining
Non-homologous end joining (NHEJ) is a pathway that repairs double-strand breaks in DNA. NHEJ is referred to as "non-homologous" because the break ends are directly ligated without the need for a homologous template, in contrast to homology direc ...
and homologous recombination
Homologous recombination is a type of genetic recombination in which genetic information is exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acids (usually DNA as in cellular organisms but may ...
respectively in many laboratory model organisms. Alongside zinc finger nuclease
Zinc-finger nucleases (ZFNs) are artificial restriction enzymes generated by fusing a zinc finger DNA-binding domain to a DNA-cleavage domain. Zinc finger domains can be engineered to target specific desired DNA sequences and this enables zin ...
s and Transcription activator-like effector nuclease
Transcription activator-like effector nucleases (TALEN) are restriction enzymes that can be engineered to cut specific sequences of DNA. They are made by fusing a TAL effector DNA-binding domain to a DNA cleavage domain (a nuclease which cuts DNA ...
(TALEN) proteins, Cas9 is becoming a prominent tool in the field of genome editing.
Cas9 has gained traction in recent years because it can cleave nearly any sequence complementary to the guide RNA. Because the target specificity of Cas9 stems from the guide RNA:DNA complementarity and not modifications to the protein itself (like TALENs and zinc fingers
A zinc finger is a small protein structural motif that is characterized by the coordination of one or more zinc ions (Zn2+) in order to stabilize the fold. It was originally coined to describe the finger-like appearance of a hypothesized struct ...
), engineering Cas9 to target new DNA is straightforward. Versions of Cas9 that bind but do not cleave cognate DNA can be used to locate transcriptional activator or repressors to specific DNA sequences in order to control transcriptional activation and repression. Native Cas9 requires a guide RNA composed of two disparate RNAs that associate – the CRISPR RNA (crRNA), and the trans-activating crRNA (tracrRNA In molecular biology, trans-activating crispr RNA (tracrRNA) is a small ''trans''-encoded RNA. It was first discovered by Emmanuelle Charpentier in her study of human pathogen ''Streptococcus pyogenes'', a type of bacteria that causes harm to human ...
). Cas9 targeting has been simplified through the engineering of a chimeric single guide RNA (chiRNA). Scientists have suggested that Cas9-based gene drive
A gene drive is a natural process and technology of genetic engineering that propagates a particular suite of genes throughout a population by altering the probability that a specific allele will be transmitted to offspring (instead of the Mende ...
s may be capable of editing the genomes of entire populations of organisms. In 2015, Cas9 was used to modify the genome of human embryos for the first time.
CRISPR-mediated immunity
To survive in a variety of challenging, inhospitable habitats that are filled withbacteriophage
A bacteriophage (), also known informally as a ''phage'' (), is a duplodnaviria virus that infects and replicates within bacteria and archaea. The term was derived from "bacteria" and the Greek φαγεῖν ('), meaning "to devour". Bacteri ...
s, bacteria and archaea have evolved methods to evade and fend off predatory virus
A virus is a submicroscopic infectious agent that replicates only inside the living cells of an organism. Viruses infect all life forms, from animals and plants to microorganisms, including bacteria and archaea.
Since Dmitri Ivanovsky's 1 ...
es. This includes the CRISPR system of adaptive immunity. In practice, CRISPR/Cas systems act as self-programmable restriction enzymes. CRISPR loci are composed of short, palindromic repeats that occur at regular intervals composed of alternate CRISPR repeats and variable CRISPR spacers between 24 and 48 nucleotides long. These CRISPR loci are usually accompanied by adjacent CRISPR-associated (cas) genes. In 2005, it was discovered by three separate groups that the spacer regions were homologous to foreign DNA elements, including plasmids and viruses. These reports provided the first biological evidence that CRISPRs might function as an immune system.
Cas9 has been used often as a genome-editing tool. Cas9 has been used in recent developments in preventing viruses from manipulating hosts' DNA. Since the CRISPR-Cas9 was developed from bacterial genome systems, it can be used to target the genetic material in viruses. The use of the enzyme Cas9 can be a solution to many viral infections. Cas9 possesses the ability to target specific viruses by the targeting of specific strands of the viral genetic information. More specifically the Cas9 enzyme targets certain sections of the viral genome that prevents the virus from carrying out its normal function. Cas9 has also been used to disrupt the detrimental strand of DNA and RNA that cause diseases and mutated strands of DNA. Cas9 has already showed promise in disrupting the effects of HIV-1. Cas9 has been shown to suppress the expression of the long terminal repeats in HIV-1. When introduced into the HIV-1 genome Cas9 has shown the ability to mutate strands of HIV-1. Cas9 has also been used in the treatment of Hepatitis B
Hepatitis B is an infectious disease caused by the ''Hepatitis B virus'' (HBV) that affects the liver; it is a type of viral hepatitis. It can cause both acute and chronic infection.
Many people have no symptoms during an initial infection. Fo ...
through targeting of the ends of certain of long terminal repeats in the Hepatitis B viral genome. Cas9 has been used to repair the mutations causing cataracts in mice.
cas3
Cas3 is an ATP-dependent single-strand DNA (ssDNA) translocase/helicase enzyme that degrades DNA as part of CRISPR based immunity.
Cas3 is a "signature" protein of class 1 CRISPR systems and functions in a complex known as CASCADE, with other c ...
, cas9, and cas10 are signature genes for type I, type II, and type III, respectively.
CRISPR-Cas defense stages
Adaptation
Adaptation
In biology, adaptation has three related meanings. Firstly, it is the dynamic evolutionary process of natural selection that fits organisms to their environment, enhancing their evolutionary fitness. Secondly, it is a state reached by the po ...
involves recognition and integration of spacers between two adjacent repeats in the CRISPR locus. The "Protospacer" refers to the sequence on the viral genome that corresponds to the spacer. A short stretch of conserved nucleotides exists proximal to the protospacer, which is called the protospacer adjacent motif (PAM). The PAM is a recognition motif that is used to acquire the DNA fragment. In type II, Cas9 recognizes the PAM during adaptation in order to ensure the acquisition of functional spacers.
Loss of spacers and even groups of several have also been observed by Aranaz et al. 2004 and Pourcel et al. 2007. This probably occurs through homologous recombination of the between-repeat material.
CRISPR processing/biogenesis
CRISPR expression includes thetranscription
Transcription refers to the process of converting sounds (voice, music etc.) into letters or musical notes, or producing a copy of something in another medium, including:
Genetics
* Transcription (biology), the copying of DNA into RNA, the fir ...
of a primary transcript called a CRISPR RNA (pre-crRNA), which is transcribed from the CRISPR locus by RNA polymerase. Specific endoribonuclease
An endoribonuclease is a ribonuclease endonuclease. It cleaves either single-stranded or double-stranded RNA, depending on the enzyme. Example includes both single proteins such as RNase III, RNase A, RNase T1, RNase T2 and RNase H and also com ...
s then cleave the pre-crRNAs into small CRISPR RNAs (crRNAs).
Interference/immunity
Interference involves the crRNAs within a multi-protein complex called CASCADE, which can recognize and specifically base-pair with regions of inserting complementary foreign DNA. The crRNA-foreign nucleic acid complex is then cleaved, however if there are mismatches between the spacer and the target DNA, or if there are mutations in the PAM, then cleavage will not be initiated. In the latter scenario, the foreign DNA is not targeted for attack by the cell, thus the replication of the virus proceeds and the host is not immune to viral infection. The interference stage can be mechanistically and temporally distinct from CRISPR acquisition and expression, yet for complete function as a defense system, all three phases must be functional. Stage 1: CRISPR spacer integration. Protospacers and protospacer-associated motifs (shown in red) are acquired at the "leader" end of a CRISPR array in the host DNA. The CRISPR array is composed of spacer sequences (shown in colored boxes) flanked by repeats (black diamonds). This process requires Cas1 and Cas2 (and Cas9 in type II), which are encoded in the cas locus, which are usually located near the CRISPR array. Stage 2: CRISPR expression. Pre-crRNA is transcribed starting at the leader region by the host RNA polymerase and then cleaved by Cas proteins into smaller crRNAs containing a single spacer and a partial repeat (shown as hairpin structure with colored spacers). Stage 3: CRISPR interference. crRNA with a spacer that has strong complementarity to the incoming foreign DNA begins a cleavage event (depicted with scissors), which requires Cas proteins. DNA cleavage interferes with viral replication and provides immunity to the host. The interference stage can be functionally and temporarily distinct from CRISPR acquisition and expression (depicted by white line dividing the cell).Transcription deactivation using dCas9
dCas9, also referred to as endonuclease deficient Cas9 can be utilized to edit gene expression when applied to the transcription binding site of the desired section of a gene. The optimal function of dCas9 is attributed to its mode of action. Gene expression is inhibited when nucleotides are no longer added to the RNA chain and therefore terminating elongation of that chain, and as a result affects the transcription process. This process occurs when dCas9 is mass-produced so it is able to affect the most genes at any given time via a sequence specific guide RNA molecule. Since dCas9 appears to down regulate gene expression, this action is amplified even more when it is used in conjunction with repressive chromatin modifier domains. The dCas9 protein has other functions outside of the regulation of gene expression. A promoter can be added to the dCas9 protein which allows them to work with each other to become efficient at beginning or stopping transcription at different sequences along a strand of DNA. These two proteins are specific in where they act on a gene. This is prevalent in certain types of prokaryotes when a promoter and dCas9 align themselves together to impede the ability of elongation of polymer of nucleotides coming together to form a transcribed piece of DNA. Without the promoter, the dCas9 protein does not have the same effect by itself or with a gene body. When examining the effects of repression of transcription further, H3K27, an amino acid component of a histone, becomes methylated through the interaction of dCas9 and a peptide called FOG1. Essentially, this interaction causes gene repression on the C + N terminal section of the amino acid complex at the specific junction of the gene, and as a result, terminates transcription. dCas9 also proves to be efficient when it comes to altering certain proteins that can create diseases. When the dCas9 attaches to a form of RNA called guide-RNA, it prevents the proliferation of repeating codons and DNA sequences that might be harmful to an organism's genome. Essentially, when multiple repeat codons are produced, it elicits a response, or recruits an abundance of dCas9 to combat the overproduction of those codons and results in the shut-down of transcription. dCas9 works synergistically with gRNA and directly affects the DNA polymerase II from continuing transcription. Further explanation of how the dCas9 protein works can be found in their utilization of plant genomes by the regulation of gene production in plants to either increase or decrease certain characteristics. The CRISPR-CAS9 system has the ability to either upregulate or downregulate genes. The dCas9 proteins are a component of the CRISPR-CAS9 system and these proteins can repress certain areas of a plant gene. This happens when dCAS9 binds to repressor domains, and in the case of the plants, deactivation of a regulatory gene such as AtCSTF64, does occur. Bacteria are another focus of the usage of dCas9 proteins as well. Since eukaryotes have a larger DNA makeup and genome; the much smaller bacteria are easy to manipulate. As a result, eukaryotes use dCas9 to inhibit RNA polymerase from continuing the process of transcription of genetic material.Structural and biochemical studies
Crystal structure
 Cas9 features a bi-lobed architecture with the guide RNA nestled between the alpha-helical lobe (blue) and the nuclease lobe (cyan, orange, and gray). These two lobes are connected through a single bridge helix. There are two nuclease domains located in the multi-domain nuclease lobe, the RuvC (gray) which cleaves the non-target DNA strand, and the HNH nuclease domain (cyan) that cleaves the target strand of DNA. The RuvC domain is encoded by sequentially disparate sites that interact in the tertiary structure to form the RuvC cleavage domain (See right figure).
Cas9 features a bi-lobed architecture with the guide RNA nestled between the alpha-helical lobe (blue) and the nuclease lobe (cyan, orange, and gray). These two lobes are connected through a single bridge helix. There are two nuclease domains located in the multi-domain nuclease lobe, the RuvC (gray) which cleaves the non-target DNA strand, and the HNH nuclease domain (cyan) that cleaves the target strand of DNA. The RuvC domain is encoded by sequentially disparate sites that interact in the tertiary structure to form the RuvC cleavage domain (See right figure).
 A key feature of the target DNA is that it must contain a protospacer adjacent motif (PAM) consisting of the three-nucleotide sequence- NGG. This PAM is recognized by the PAM-interacting domain (PI domain, orange) located near the C-terminal end of Cas9. Cas9 undergoes distinct conformational changes between the apo, guide RNA bound, and guide RNA:DNA bound states.
Cas9 recognizes the
A key feature of the target DNA is that it must contain a protospacer adjacent motif (PAM) consisting of the three-nucleotide sequence- NGG. This PAM is recognized by the PAM-interacting domain (PI domain, orange) located near the C-terminal end of Cas9. Cas9 undergoes distinct conformational changes between the apo, guide RNA bound, and guide RNA:DNA bound states.
Cas9 recognizes the stem-loop
Stem-loop intramolecular base pairing is a pattern that can occur in single-stranded RNA. The structure is also known as a hairpin or hairpin loop. It occurs when two regions of the same strand, usually complementary in nucleotide sequence when ...
architecture inherent in the CRISPR locus, which mediates the maturation of crRNA-tracrRNA ribonucleoprotein
Nucleoproteins are proteins conjugated with nucleic acids (either DNA or RNA). Typical nucleoproteins include ribosomes, nucleosomes and viral nucleocapsid proteins.
Structures
Nucleoproteins tend to be positively charged, facilitating in ...
complex. Cas9 in complex with CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA) further recognizes and degrades the target dsDNA. In the co-crystal structure shown here, the crRNA-tracrRNA complex is replaced by a chimeric single-guide RNA (sgRNA, in red) which has been proved to have the same function as the natural RNA complex. The sgRNA base paired with target ssDNA is anchored by Cas9 as a T-shaped architecture. This crystal structure of the DNA-bound Cas9 enzyme reveals distinct conformational changes in the alpha-helical lobe with respect to the nuclease lobe, as well as the location of the HNH domain. The protein consists of a recognition lobe (REC) and a nuclease lobe (NUC). All regions except the HNH form tight interactions with each other and sgRNA-ssDNA complex, while the HNH domain forms few contacts with the rest of the protein. In another conformation of Cas9 complex observed in the crystal, the HNH domain is not visible. These structures suggest the conformational flexibility of HNH domain.
To date, at least three crystal structures have been studied and published. One representing a conformation of Cas9 in the apo state, and two representing Cas9 in the DNA bound state.
Interactions with sgRNA
DNA cleavage
cofactors
Cofactor may also refer to:
* Cofactor (biochemistry), a substance that needs to be present in addition to an enzyme for a certain reaction to be catalysed
* A domain parameter in elliptic curve cryptography, defined as the ratio between the order ...
for the RNA-mediated DNA cleavage; however, Cas9 has been shown to exhibit varying levels of activity in the presence of other divalent
In chemistry, the valence (US spelling) or valency (British spelling) of an element is the measure of its combining capacity with other atoms when it forms chemical compounds or molecules.
Description
The combining capacity, or affinity of an ...
metal ions. For instance, Cas9 in the presence of manganese (Mn2+) has been shown to be capable of RNA-independent DNA cleavage. The kinetics
Kinetics ( grc, κίνησις, , kinesis, ''movement'' or ''to move'') may refer to:
Science and medicine
* Kinetics (physics), the study of motion and its causes
** Rigid body kinetics, the study of the motion of rigid bodies
* Chemical ki ...
of DNA cleavage by Cas9 have been of great interest to the scientific community, as this data provides insight into the intricacies of the reaction. While the cleavage of DNA by RNA-bound Cas9 has been shown to be relatively rapid ('' k'' ≥ 700 s−1), the release of the cleavage products is very slow ( ''t''1/2 = ln(2)/''k'' ≈ 43-91 h), essentially rendering Cas9 a single- turnover enzyme. Additional studies regarding the kinetics of Cas9 have shown engineered Cas9 to be effective in reducing off-target effects by modifying the rate of the reaction.
The cleavage efficiency of Cas9 depends on numerous factors. A key requirement is the presence of a valid PAM at the non-target strand 3 nucleotides downstream from the cleavage site. The canonical PAM sequence for ''S. Pyogenes'' Cas9 is NGG, but alternative motifs are tolerated with lower cleavage activity. The most efficient alternative PAM motifs for the wild-type ''S. Pyogenes'' Cas9 are NAG and NGA. The sequence composition at the target DNA site complementary to the 20 nucletode spacer region of the gRNA also affects cleavage efficiency. The most relevant nucleotide composition properties that impact efficiency are those in the PAM-proximal region. Free energy changes of nucleic acids are also highly relevant in defining cleavage activity. Guide RNAs that bind to the DNA forming a duplex that falls into a restricted range of binding free energy changes that excludes extremely weak or stable bindings generally perform efficiently. Stable guide RNA folding conformations can also impair cleavage.
Problems bacteria pose to Cas9 editing
Most archaea and bacteria stubbornly refuse to allow a Cas9 to edit their genome. This is because they can attach foreign DNA, that does not affect them, into their genome. Another way that these cells defy Cas9 is by process of restriction modification (RM) system. When a bacteriophage enters a bacteria or archaea cell it is targeted by the RM system. The RM system then cuts the bacteriophages DNA into separate pieces by restriction enzymes and uses endonucleases to further destroy the strands of DNA. This poses a problem to Cas9 editing because the RM system also targets the foreign genes added by the Cas9 process.Applications of Cas9 to transcription tuning
Interference of transcription by dCas9
Due to the unique ability of Cas9 to bind to essentially any complement sequence in anygenome
In the fields of molecular biology and genetics, a genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding ge ...
, researchers wanted to use this enzyme to repress transcription
Transcription refers to the process of converting sounds (voice, music etc.) into letters or musical notes, or producing a copy of something in another medium, including:
Genetics
* Transcription (biology), the copying of DNA into RNA, the fir ...
of various genomic loci. To accomplish this, the two crucial catalytic residues of the RuvC and HNH domain can be mutated to alanine
Alanine (symbol Ala or A), or α-alanine, is an α-amino acid that is used in the biosynthesis of proteins. It contains an amine group and a carboxylic acid group, both attached to the central carbon atom which also carries a methyl group side ...
abolishing all endonuclease activity of Cas9. The resulting protein coined 'dead' Cas9 or 'dCas9' for short, can still tightly bind to dsDNA. This catalytic
Catalysis () is the process of increasing the rate of a chemical reaction by adding a substance known as a catalyst (). Catalysts are not consumed in the reaction and remain unchanged after it. If the reaction is rapid and the catalyst recyc ...
ally inactive Cas9 variant has been used for both mechanistic studies into Cas9 DNA interrogative binding and as a general programmable DNA binding RNA-Protein complex.
The interaction of dCas9 with target dsDNA is so tight that high molarity
Molar concentration (also called molarity, amount concentration or substance concentration) is a measure of the concentration of a chemical species, in particular of a solute in a solution, in terms of amount of substance per unit volume of solu ...
urea
Urea, also known as carbamide, is an organic compound with chemical formula . This amide has two amino groups (–) joined by a carbonyl functional group (–C(=O)–). It is thus the simplest amide of carbamic acid.
Urea serves an important r ...
protein denaturant can not fully dissociate the dCas9 RNA-protein complex from dsDNA target. dCas9 has been targeted with engineered single guide RNAs to transcription initiation sites of any loci where dCas9 can compete with RNA polymerase at promoters to halt transcription. Also, dCas9 can be targeted to the coding region of loci such that inhibition of RNA Polymerase occurs during the elongation phase of transcription. In Eukaryotes, silencing of gene expression can be extended by targeting dCas9 to enhancer sequences, where dCas9 can block assembly of transcription factors leading to silencing of specific gene expression. Moreover, the guide RNAs provided to dCas9 can be designed to include specific mismatches to its complementary cognate sequence that will quantitatively weaken the interaction of dCas9 for its programmed cognate sequence allowing a researcher to tune the extent of gene silencing applied to a gene of interest.
This technology is similar in principle to RNAi
RNA interference (RNAi) is a biological process in which RNA molecules are involved in sequence-specific suppression of gene expression by double-stranded RNA, through translational or transcriptional repression. Historically, RNAi was known by ...
such that gene expression is being modulated at the RNA level. However, the dCas9 approach has gained much traction as there exist less off-target effects and in general larger and more reproducible silencing effects through the use of dCas9 compared to RNAi screens. Furthermore, because the dCas9 approach to gene silencing can be quantitatively controlled, a researcher can now precisely control the extent to which a gene of interest is repressed allowing more questions about gene regulation and gene stoichiometry to be answered.
Beyond direct binding of dCas9 to transcriptionally sensitive positions of loci, dCas9 can be fused to a variety of modulatory protein domains to carry out a myriad of functions. Recently, dCas9 has been fused to chromatin
Chromatin is a complex of DNA and protein found in eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important roles in r ...
remodeling proteins (HDACs/HATs) to reorganize the chromatin structure around various loci. This is important in targeting various eukaryotic genes of interest as heterochromatin structures hinder Cas9 binding. Furthermore, because Cas9 can react to heterochromatin
Heterochromatin is a tightly packed form of DNA or '' condensed DNA'', which comes in multiple varieties. These varieties lie on a continue between the two extremes of constitutive heterochromatin and facultative heterochromatin. Both play a role ...
, it is theorized that this enzyme can be further applied to studying the chromatin structure of various loci. Additionally, dCas9 has been employed in genome wide screens of gene repression. By employing large libraries of guide RNAs capable of targeting thousands of genes, genome wide genetic screens using dCas9 have been conducted.
Another method for silencing transcription with Cas9 is to directly cleave mRNA products with the catalytically active Cas9 enzyme. This approach is made possible by hybridizing ssDNA with a PAM complement sequence to ssRNA allowing for a dsDNA-RNA PAM site for Cas9 binding. This technology makes available the ability to isolate endogenous RNA transcripts in cells without the need to induce chemical modifications to RNA or RNA tagging methods.
Transcription activation by dCas9 fusion proteins
In contrast to silencing genes, dCas9 can also be used to activate genes when fused to transcription activating factors. These factors include subunits of bacterial RNA Polymerase II and traditional transcription factors in eukaryotes. Recently, genome-wide screens of transcription activation have also been accomplished using dCas9 fusions named 'CRISPRa' for activation.See also
* DCas9 activation system * CRISPR *CRISPR gene editing
CRISPR gene editing (pronounced "crisper") is a genetic engineering technique in molecular biology by which the genomes of living organisms may be modified. It is based on a simplified version of the bacterial CRISPR-Cas9 antiviral defense sys ...
* Genome editing
Genome editing, or genome engineering, or gene editing, is a type of genetic engineering in which DNA is inserted, deleted, modified or replaced in the genome of a living organism. Unlike early genetic engineering techniques that randomly inserts ...
* Zinc finger nuclease
Zinc-finger nucleases (ZFNs) are artificial restriction enzymes generated by fusing a zinc finger DNA-binding domain to a DNA-cleavage domain. Zinc finger domains can be engineered to target specific desired DNA sequences and this enables zin ...
* Transcription activator-like effector nuclease
Transcription activator-like effector nucleases (TALEN) are restriction enzymes that can be engineered to cut specific sequences of DNA. They are made by fusing a TAL effector DNA-binding domain to a DNA cleavage domain (a nuclease which cuts DNA ...
References
Further reading
* * **External links
* {{Authority control Enzymes Genetic engineering Repetitive DNA sequences Immune system Bacterial proteins Genome editing