Complex V on:
[Wikipedia]
[Google]
[Amazon]
ATP synthase is an
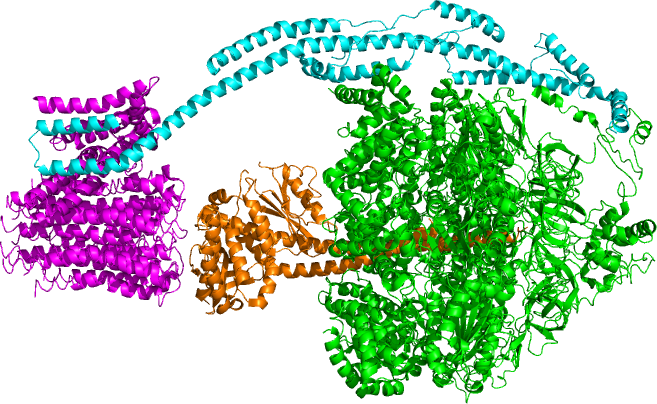 Located within the
Located within the
 FO is a water
FO is a water
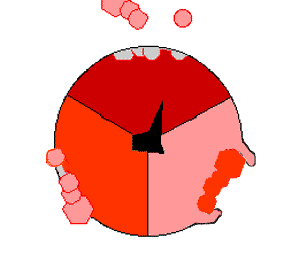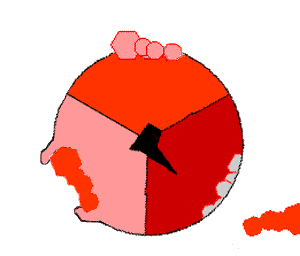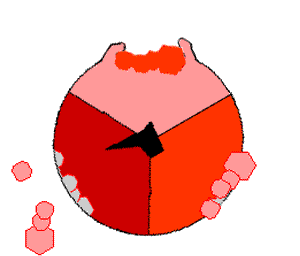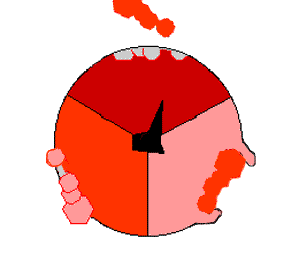 In the 1960s through the 1970s, Paul Boyer, a
In the 1960s through the 1970s, Paul Boyer, a
''The Vital Question: Energy, Evolution, and the Origins of Complex Life''
Ww Norton, 2015-07-20, (Link points to Figure 10 showing model of ATP synthase)
"ATP synthase — a splendid molecular machine"
* Well illustrate
by Antony Crofts of the
Proton and Sodium translocating F-type, V-type and A-type ATPases in OPM database
to Paul D. Boyer and John E. Walker for the enzymatic mechanism of synthesis of ATP; and to Jens C. Skou, for discovery of an ion-transporting enzyme, , -ATPase.
– ATP synthesis animation * David Goodsell
"ATP Synthase- Molecule of the Month"
{{Portal bar, Biology, border=no Enzymes Cellular respiration Photosynthesis EC 3.6.3 Integral membrane proteins Protein complexes
enzyme
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different mol ...
that catalyzes the formation of the energy storage molecule adenosine triphosphate
Adenosine triphosphate (ATP) is a nucleoside triphosphate that provides energy to drive and support many processes in living cell (biology), cells, such as muscle contraction, nerve impulse propagation, and chemical synthesis. Found in all known ...
(ATP) using adenosine diphosphate
Adenosine diphosphate (ADP), also known as adenosine pyrophosphate (APP), is an important organic compound in metabolism and is essential to the flow of energy in living cells. ADP consists of three important structural components: a sugar backbon ...
(ADP) and inorganic phosphate
Phosphates are the naturally occurring form of the element phosphorus.
In chemistry, a phosphate is an anion, salt, functional group or ester derived from a phosphoric acid. It most commonly means orthophosphate, a derivative of orthop ...
(Pi). ATP synthase is a molecular machine
Molecular machines are a class of molecules typically described as an assembly of a discrete number of molecular components intended to produce mechanical movements in response to specific stimuli, mimicking macromolecular devices such as switch ...
. The overall reaction catalyzed by ATP synthase is:
* ADP + Pi + 2H+out ATP + H2O + 2H+in
ATP synthase lies across a cellular membrane and forms an aperture that protons
A proton is a stable subatomic particle, symbol , H+, or 1H+ with a positive electric charge of +1 ''e'' ( elementary charge). Its mass is slightly less than the mass of a neutron and approximately times the mass of an electron (the pro ...
can cross from areas of high concentration to areas of low concentration, imparting energy for the synthesis of ATP. This electrochemical gradient
An electrochemical gradient is a gradient of electrochemical potential, usually for an ion that can move across a membrane. The gradient consists of two parts:
* The chemical gradient, or difference in Concentration, solute concentration across ...
is generated by the electron transport chain
An electron transport chain (ETC) is a series of protein complexes and other molecules which transfer electrons from electron donors to electron acceptors via redox reactions (both reduction and oxidation occurring simultaneously) and couples th ...
and allows cells to store energy in ATP for later use. In prokaryotic cells ATP synthase lies across the plasma membrane
The cell membrane (also known as the plasma membrane or cytoplasmic membrane, and historically referred to as the plasmalemma) is a biological membrane that separates and protects the interior of a cell from the outside environment (the extr ...
, while in eukaryotic cells
The eukaryotes ( ) constitute the domain of Eukaryota or Eukarya, organisms whose cells have a membrane-bound nucleus. All animals, plants, fungi, seaweeds, and many unicellular organisms are eukaryotes. They constitute a major group of li ...
it lies across the inner mitochondrial membrane
The inner mitochondrial membrane (IMM) is the mitochondrial membrane which separates the mitochondrial matrix from the intermembrane space.
Structure
The structure of the inner mitochondrial membrane is extensively folded and compartmentalized. T ...
. Organisms capable of photosynthesis
Photosynthesis ( ) is a system of biological processes by which photosynthetic organisms, such as most plants, algae, and cyanobacteria, convert light energy, typically from sunlight, into the chemical energy necessary to fuel their metabo ...
also have ATP synthase across the thylakoid membrane
Thylakoids are membrane-bound compartments inside chloroplasts and cyanobacteria. They are the site of the light-dependent reactions of photosynthesis. Thylakoids consist of a thylakoid membrane surrounding a thylakoid lumen. Chloroplast thyl ...
, which in plants
Plants are the eukaryotes that form the kingdom Plantae; they are predominantly photosynthetic. This means that they obtain their energy from sunlight, using chloroplasts derived from endosymbiosis with cyanobacteria to produce sugars f ...
is located in the chloroplast
A chloroplast () is a type of membrane-bound organelle, organelle known as a plastid that conducts photosynthesis mostly in plant cell, plant and algae, algal cells. Chloroplasts have a high concentration of chlorophyll pigments which captur ...
and in cyanobacteria
Cyanobacteria ( ) are a group of autotrophic gram-negative bacteria that can obtain biological energy via oxygenic photosynthesis. The name "cyanobacteria" () refers to their bluish green (cyan) color, which forms the basis of cyanobacteri ...
is located in the cytoplasm
The cytoplasm describes all the material within a eukaryotic or prokaryotic cell, enclosed by the cell membrane, including the organelles and excluding the nucleus in eukaryotic cells. The material inside the nucleus of a eukaryotic cell a ...
.
Eukaryotic ATP synthases are F-ATPase
F-ATPase, also known as F-Type ATPase, is an ATPase/ synthase found in bacterial plasma membranes, in mitochondrial inner membranes (in oxidative phosphorylation, where it is known as Complex V), and in chloroplast thylakoid membranes. It us ...
s (which usually work as ATP synthases instead of ATPases in cellular environments) and running "in reverse" for an ATPase
ATPases (, Adenosine 5'-TriPhosphatase, adenylpyrophosphatase, ATP monophosphatase, triphosphatase, ATP hydrolase, adenosine triphosphatase) are a class of enzymes that catalyze the decomposition of ATP into ADP and a free phosphate ion or ...
(ATPase catalyze
Catalysis () is the increase in rate of a chemical reaction due to an added substance known as a catalyst (). Catalysts are not consumed by the reaction and remain unchanged after it. If the reaction is rapid and the catalyst recycles quick ...
the decomposition
Decomposition is the process by which dead organic substances are broken down into simpler organic or inorganic matter such as carbon dioxide, water, simple sugars and mineral salts. The process is a part of the nutrient cycle and is ess ...
of ATP into ADP and a free phosphate ion). This article deals mainly with this type. An F-ATPase consists of two main subunits, FO and F1, which has a rotational motor mechanism allowing for ATP production.
Nomenclature
The F1 fraction derives its name from the term "Fraction 1" and FO (written as a subscript letter "o", not "zero") derives its name from being the binding fraction foroligomycin
Oligomycins are macrolides created by ''Streptomyces'' that are strong antibacterial agents but are often poisonous to other organisms, including humans.
Function
Oligomycins have use as antibiotics. However, in humans, they have limited or n ...
, a type of naturally derived antibiotic that is able to inhibit the FO unit of ATP synthase. These functional regions consist of different protein subunits — refer to tables. This enzyme is used in synthesis of ATP through aerobic respiration.
Structure and function
 Located within the
Located within the thylakoid membrane
Thylakoids are membrane-bound compartments inside chloroplasts and cyanobacteria. They are the site of the light-dependent reactions of photosynthesis. Thylakoids consist of a thylakoid membrane surrounding a thylakoid lumen. Chloroplast thyl ...
and the inner mitochondrial membrane
The inner mitochondrial membrane (IMM) is the mitochondrial membrane which separates the mitochondrial matrix from the intermembrane space.
Structure
The structure of the inner mitochondrial membrane is extensively folded and compartmentalized. T ...
, ATP synthase consists of two regions FO and F1. FO causes rotation of F1 and is made of c-ring and subunits a, two b, F6. F1 is made of α, β, γ, and δ subunits. F1 has a water-soluble part that can hydrolyze ATP. FO on the other hand has mainly hydrophobic regions. FO F1 creates a pathway for protons movement across the membrane.
F1 region
The F1 portion of ATP synthase ishydrophilic
A hydrophile is a molecule or other molecular entity that is attracted to water molecules and tends to be dissolved by water.Liddell, H.G. & Scott, R. (1940). ''A Greek-English Lexicon'' Oxford: Clarendon Press.
In contrast, hydrophobes are n ...
and responsible for hydrolyzing ATP. The F1 unit protrudes into the mitochondrial matrix
In the mitochondrion, the matrix is the space within the inner membrane. It can also be referred as the mitochondrial fluid. The word "matrix" stems from the fact that this space is viscous, compared to the relatively aqueous cytoplasm. The mitoc ...
space. Subunits α and β make a hexamer with 6 binding sites. Three of them are catalytically inactive and they bind ADP.
Three other subunits catalyze the ATP synthesis. The other F1 subunits γ, δ, and ε are a part of a rotational motor mechanism (rotor/axle). The γ subunit allows β to go through conformational changes (i.e., closed, half open, and open states) that allow for ATP to be bound and released once synthesized. The F1 particle is large and can be seen in the transmission electron microscope by negative staining. These are particles of 9 nm diameter that pepper the inner mitochondrial membrane.
FO region
 FO is a water
FO is a water insoluble
In chemistry, solubility is the ability of a substance, the solute, to form a solution with another substance, the solvent. Insolubility is the opposite property, the inability of the solute to form such a solution.
The extent of the solub ...
protein with eight subunits and a transmembrane ring. The ring has a tetramer
A tetramer () (''tetra-'', "four" + '' -mer'', "parts") is an oligomer formed from four monomers or subunits. The associated property is called ''tetramery''. An example from inorganic chemistry is titanium methoxide with the empirical formula ...
ic shape with a helix-loop-helix protein that goes through conformational changes when protonated and deprotonated, pushing neighboring subunits to rotate, causing the spinning of FO which then also affects conformation of F1, resulting in switching of states of alpha and beta subunits. The FO region of ATP synthase is a proton pore that is embedded in the mitochondrial membrane. It consists of three main subunits, a, b, and c. Six c subunits make up the rotor ring, and subunit b makes up a stalk connecting to F1 OSCP that prevents the αβ hexamer from rotating. Subunit a connects b to the c ring. Humans have six additional subunits, d, e, f, g, F6, and 8 (or A6L). This part of the enzyme is located in the mitochondrial inner membrane and couples proton translocation to the rotation that causes ATP synthesis in the F1 region.
In eukaryotes, mitochondrial FO forms membrane-bending dimers. These dimers self-arrange into long rows at the end of the cristae
A crista (; : cristae) is a fold in the inner membrane of a mitochondrion. The name is from the Latin for ''crest'' or ''plume'', and it gives the inner membrane its characteristic wrinkled shape, providing a large amount of surface area for che ...
, possibly the first step of cristae formation. An atomic model for the dimeric yeast FO region was determined by cryo-EM at an overall resolution of 3.6 Å.
Binding model
 In the 1960s through the 1970s, Paul Boyer, a
In the 1960s through the 1970s, Paul Boyer, a UCLA
The University of California, Los Angeles (UCLA) is a public land-grant research university in Los Angeles, California, United States. Its academic roots were established in 1881 as a normal school then known as the southern branch of the C ...
Professor, developed the binding change, or flip-flop, mechanism theory, which postulated that ATP synthesis is dependent on a conformational change in ATP synthase generated by rotation of the gamma subunit. The research group of John E. Walker, then at the MRC Laboratory of Molecular Biology
The Medical Research Council (MRC) Laboratory of Molecular Biology (LMB) is a research institute in Cambridge, England, involved in the revolution in molecular biology which occurred in the 1950–60s. Since then it has remained a major medical r ...
in Cambridge
Cambridge ( ) is a List of cities in the United Kingdom, city and non-metropolitan district in the county of Cambridgeshire, England. It is the county town of Cambridgeshire and is located on the River Cam, north of London. As of the 2021 Unit ...
, crystallized the F1 catalytic-domain of ATP synthase. The structure, at the time the largest asymmetric protein structure known, indicated that Boyer's rotary-catalysis model was, in essence, correct. For elucidating this, Boyer and Walker shared half of the 1997 Nobel Prize in Chemistry
The Nobel Prize in Chemistry () is awarded annually by the Royal Swedish Academy of Sciences to scientists in the various fields of chemistry. It is one of the five Nobel Prizes established by the will of Alfred Nobel in 1895, awarded for outst ...
.
The crystal structure of the F1 showed alternating alpha and beta subunits (3 of each), arranged like segments of an orange around a rotating asymmetrical gamma subunit. According to the current model of ATP synthesis (known as the alternating catalytic model), the transmembrane potential created by (H+) proton cations supplied by the electron transport chain, drives the (H+) proton cations from the intermembrane space through the membrane via the FO region of ATP synthase. A portion of the FO (the ring of c-subunits) rotates as the protons pass through the membrane. The c-ring is tightly attached to the asymmetric central stalk (consisting primarily of the gamma subunit), causing it to rotate within the alpha3beta3 of F1 causing the 3 catalytic nucleotide binding sites to go through a series of conformational changes that lead to ATP synthesis. The major F1 subunits are prevented from rotating in sympathy with the central stalk rotor by a peripheral stalk that joins the alpha3beta3 to the non-rotating portion of FO. The structure of the intact ATP synthase is currently known at low-resolution from electron cryo-microscopy (cryo-EM) studies of the complex. The cryo-EM model of ATP synthase suggests that the peripheral stalk is a flexible structure that wraps around the complex as it joins F1 to FO. Under the right conditions, the enzyme reaction can also be carried out in reverse, with ATP hydrolysis driving proton pump
A proton pump is an integral membrane protein pump that builds up a proton gradient across a biological membrane. Proton pumps catalyze the following reaction:
: n one side of a biological membrane/sub> + energy n the other side of the m ...
ing across the membrane.
The binding change mechanism involves the active site of a β subunit's cycling between three states. In the "loose" state, ADP and phosphate enter the active site; in the adjacent diagram, this is shown in pink. The enzyme then undergoes a change in shape and forces these molecules together, with the active site in the resulting "tight" state (shown in red) binding the newly produced ATP molecule with very high affinity
Affinity may refer to:
Commerce, finance and law
* Affinity (law), kinship by marriage
* Affinity analysis, a market research and business management technique
* Affinity Credit Union, a Saskatchewan-based credit union
* Affinity Equity Pa ...
. Finally, the active site cycles back to the open state (orange), releasing ATP and binding more ADP and phosphate, ready for the next cycle of ATP production.
Physiological role
Like other enzymes, the activity of F1FO ATP synthase is reversible. Large-enough quantities of ATP cause it to create a transmembraneproton
A proton is a stable subatomic particle, symbol , Hydron (chemistry), H+, or 1H+ with a positive electric charge of +1 ''e'' (elementary charge). Its mass is slightly less than the mass of a neutron and approximately times the mass of an e ...
gradient
In vector calculus, the gradient of a scalar-valued differentiable function f of several variables is the vector field (or vector-valued function) \nabla f whose value at a point p gives the direction and the rate of fastest increase. The g ...
, this is used by fermenting bacteria that do not have an electron transport chain, but rather hydrolyze ATP to make a proton gradient, which they use to drive flagella
A flagellum (; : flagella) (Latin for 'whip' or 'scourge') is a hair-like appendage that protrudes from certain plant and animal sperm cells, from fungal spores ( zoospores), and from a wide range of microorganisms to provide motility. Many pr ...
and the transport of nutrients into the cell.
In respiring bacteria
Bacteria (; : bacterium) are ubiquitous, mostly free-living organisms often consisting of one Cell (biology), biological cell. They constitute a large domain (biology), domain of Prokaryote, prokaryotic microorganisms. Typically a few micr ...
under physiological conditions, ATP synthase, in general, runs in the opposite direction, creating ATP while using the proton motive force
Chemiosmosis is the movement of ions across a semipermeable membrane bound structure, down their electrochemical gradient. An important example is the formation of adenosine triphosphate (ATP) by the movement of hydrogen ions (H+) across a membra ...
created by the electron transport chain
An electron transport chain (ETC) is a series of protein complexes and other molecules which transfer electrons from electron donors to electron acceptors via redox reactions (both reduction and oxidation occurring simultaneously) and couples th ...
as a source of energy. The overall process of creating energy in this fashion is termed oxidative phosphorylation
Oxidative phosphorylation(UK , US : or electron transport-linked phosphorylation or terminal oxidation, is the metabolic pathway in which Cell (biology), cells use enzymes to Redox, oxidize nutrients, thereby releasing chemical energy in order ...
.
The same process takes place in the mitochondria
A mitochondrion () is an organelle found in the cells of most eukaryotes, such as animals, plants and fungi. Mitochondria have a double membrane structure and use aerobic respiration to generate adenosine triphosphate (ATP), which is us ...
, where ATP synthase is located in the inner mitochondrial membrane and the F1-part projects into the mitochondrial matrix
In the mitochondrion, the matrix is the space within the inner membrane. It can also be referred as the mitochondrial fluid. The word "matrix" stems from the fact that this space is viscous, compared to the relatively aqueous cytoplasm. The mitoc ...
. By pumping proton cations into the matrix, the ATP-synthase converts ADP into ATP.
Evolution
Theevolution
Evolution is the change in the heritable Phenotypic trait, characteristics of biological populations over successive generations. It occurs when evolutionary processes such as natural selection and genetic drift act on genetic variation, re ...
of ATP synthase is thought to have been modular whereby two functionally independent subunits became associated and gained new functionality. This association appears to have occurred early in evolutionary history, because essentially the same structure and activity of ATP synthase enzymes are present in all kingdoms of life. The F-ATP synthase displays high functional and mechanistic similarity to the V-ATPase
Vacuolar-type ATPase (V-ATPase) is a highly conserved evolutionarily ancient enzyme with remarkably diverse functions in eukaryotic organisms. V-ATPases acidify a wide array of intracellular organelles and pump protons across the plasma ...
. However, whereas the F-ATP synthase generates ATP by utilising a proton gradient, the V-ATPase
Vacuolar-type ATPase (V-ATPase) is a highly conserved evolutionarily ancient enzyme with remarkably diverse functions in eukaryotic organisms. V-ATPases acidify a wide array of intracellular organelles and pump protons across the plasma ...
generates a proton gradient at the expense of ATP, generating pH values of as low as 1.
The F1 region also shows significant similarity to hexameric DNA helicases (especially the Rho factor
A ρ factor (Rho factor) is a bacterial protein involved in the termination of transcription.
* Rho factor binds to the transcription terminator pause site, an exposed region of single stranded RNA (a stretch of 72 nucleotides) after the open ...
), and the entire enzyme region shows some similarity to -powered T3SS or flagellar motor complexes. The α3β3 hexamer of the F1 region shows significant structural similarity to hexameric DNA helicases; both form a ring with 3-fold rotational symmetry with a central pore. Both have roles dependent on the relative rotation of a macromolecule within the pore; the DNA helicases use the helical shape of DNA to drive their motion along the DNA molecule and to detect supercoiling, whereas the α3β3 hexamer uses the conformational changes through the rotation of the γ subunit to drive an enzymatic reaction.
The motor of the FO particle shows great functional similarity to the motors that drive flagella. Both feature a ring of many small alpha-helical proteins that rotate relative to nearby stationary proteins, using a potential gradient as an energy source. This link is tenuous, however, as the overall structure of flagellar motors is far more complex than that of the FO particle and the ring with about 30 rotating proteins is far larger than the 10, 11, or 14 helical proteins in the FO complex. More recent structural data do however show that the ring and the stalk are structurally similar to the F1 particle.
The modular evolution theory for the origin of ATP synthase suggests that two subunits with independent function, a DNA helicase with ATPase activity and a motor, were able to bind, and the rotation of the motor drove the ATPase activity of the helicase in reverse. This complex then evolved greater efficiency and eventually developed into today's intricate ATP synthases. Alternatively, the DNA helicase/ motor complex may have had pump activity with the ATPase activity of the helicase driving the motor in reverse. This may have evolved to carry out the reverse reaction and act as an ATP synthase.
Inhibitors
A variety of natural and synthetic inhibitors of ATP synthase have been discovered. These have been used to probe the structure and mechanism of ATP synthase. Some may be of therapeutic use. There are several classes of ATP synthase inhibitors, including peptide inhibitors, polyphenolic phytochemicals, polyketides, organotin compounds, polyenic α-pyrone derivatives, cationic inhibitors, substrate analogs, amino acid modifiers, and other miscellaneous chemicals. Some of the most commonly used ATP synthase inhibitors areoligomycin
Oligomycins are macrolides created by ''Streptomyces'' that are strong antibacterial agents but are often poisonous to other organisms, including humans.
Function
Oligomycins have use as antibiotics. However, in humans, they have limited or n ...
and DCCD.
In different organisms
Bacteria
' ATP synthase is the simplest known form of ATP synthase, with 8 different subunit types. Bacterial F-ATPases can occasionally operate in reverse, turning them into an ATPase. Some bacteria have no F-ATPase, using an A/V-type ATPase bidirectionally.Yeast
Yeast ATP synthase is one of the best-studied eukaryotic ATP synthases; and five F1, eight FO subunits, and seven associated proteins have been identified. Most of these proteins have homologues in other eukaryotes.Plant
In plants, ATP synthase is also present inchloroplasts
A chloroplast () is a type of membrane-bound organelle, organelle known as a plastid that conducts photosynthesis mostly in plant cell, plant and algae, algal cells. Chloroplasts have a high concentration of chlorophyll pigments which captur ...
(CF1FO-ATP synthase). The enzyme is integrated into thylakoid
Thylakoids are membrane-bound compartments inside chloroplasts and cyanobacterium, cyanobacteria. They are the site of the light-dependent reactions of photosynthesis. Thylakoids consist of a #Membrane, thylakoid membrane surrounding a #Lumen, ...
membrane; the CF1-part sticks into stroma, where dark reactions of photosynthesis (also called the light-independent reactions or the Calvin cycle
The Calvin cycle, light-independent reactions, bio synthetic phase, dark reactions, or photosynthetic carbon reduction (PCR) cycle of photosynthesis is a series of chemical reactions that convert carbon dioxide and hydrogen-carrier compounds into ...
) and ATP synthesis take place. The overall structure and the catalytic mechanism of the chloroplast ATP synthase are almost the same as those of the bacterial enzyme. However, in chloroplasts, the proton motive force
Chemiosmosis is the movement of ions across a semipermeable membrane bound structure, down their electrochemical gradient. An important example is the formation of adenosine triphosphate (ATP) by the movement of hydrogen ions (H+) across a membra ...
is generated not by respiratory electron transport chain but by primary photosynthetic proteins. The synthase has a 40-aa insert in the gamma-subunit to inhibit wasteful activity when dark.
Mammal
The ATP synthase isolated from bovine (''Bos taurus'') heart mitochondria is, in terms of biochemistry and structure, the best-characterized ATP synthase. Beef heart is used as a source for the enzyme because of the high concentration of mitochondria in cardiac muscle. Their genes have close homology to human ATP synthases. Human genes that encode components of ATP synthases: * '' ATP5A1'' * '' ATP5B'' * '' ATP5C1'', '' ATP5D'', '' ATP5E'', '' ATP5F1'', '' ATP5MC1'', '' ATP5G2'', '' ATP5G3'', '' ATP5H'', '' ATP5I'', '' ATP5J'', '' ATP5J2'', '' ATP5L'', '' ATP5O'' * ''MT-ATP6
''MT-ATP6'' (or ''ATP6'') is a mitochondrial gene with the full name 'mitochondrially encoded ATP synthase membrane subunit 6' that encodes the ATP synthase Fo subunit 6 (or subunit/chain A). This subunit belongs to the Fo complex of the large ...
'', ''MT-ATP8
''MT-ATP8'' (or ''ATP8'') is a mitochondrial gene with the full name 'mitochondrially encoded ATP synthase membrane subunit 8' that encodes a subunit of mitochondrial ATP synthase, ATP synthase Fo subunit 8 (or subunit A6L). This subunit be ...
''
Other eukaryotes
Eukaryotes belonging to some divergent lineages have very special organizations of the ATP synthase. Aeuglenozoa
Euglenozoa are a large group of flagellate Discoba. They include a variety of common free-living species, as well as a few important parasites, some of which infect humans. Euglenozoa are represented by four major groups, ''i.e.,'' Kinetoplastea, ...
ATP synthase forms a dimer with a boomerang-shaped F1 head like other mitochondrial ATP synthases, but the FO subcomplex has many unique subunits. It uses cardiolipin
Cardiolipin (IUPAC name 1,3-bis(''sn''-3’-phosphatidyl)-''sn''-glycerol, "''sn''" designating stereospecific numbering) is an important component of the inner mitochondrial membrane, where it constitutes about 20% of the total lipid composition. ...
. The inhibitory IF1 also binds differently, in a way shared with trypanosomatida
Trypanosomatida is a group of kinetoplastid unicellular organisms distinguished by having only a single flagellum. The name is derived from the Greek ''trypano'' (borer) and ''soma'' (body) because of the corkscrew-like motion of some trypanosom ...
.
Archaea
Archaea do not generally have an F-ATPase. Instead, they synthesize ATP using the A-ATPase/synthase, a rotary machine structurally similar to theV-ATPase
Vacuolar-type ATPase (V-ATPase) is a highly conserved evolutionarily ancient enzyme with remarkably diverse functions in eukaryotic organisms. V-ATPases acidify a wide array of intracellular organelles and pump protons across the plasma ...
but mainly functioning as an ATP synthase. Like the bacteria F-ATPase, it is believed to also function as an ATPase.
LUCA and earlier
F-ATPase gene linkage and gene order are widely conserved across ancient prokaryote lineages, implying that this system already existed at a date before thelast universal common ancestor
The last universal common ancestor (LUCA) is the hypothesized common ancestral cell from which the three domains of life, the Bacteria, the Archaea, and the Eukarya originated. The cell had a lipid bilayer; it possessed the genetic code a ...
, the LUCA.
See also
* ATP10 protein required for the assembly of the FO sector of the mitochondrial ATPase complex. *Chloroplast
A chloroplast () is a type of membrane-bound organelle, organelle known as a plastid that conducts photosynthesis mostly in plant cell, plant and algae, algal cells. Chloroplasts have a high concentration of chlorophyll pigments which captur ...
* Electron transfer chain
An electron transport chain (ETC) is a series of protein complexes and other molecules which transfer electrons from electron donors to electron acceptors via redox reactions (both reduction and oxidation occurring simultaneously) and couples this ...
* Flavoprotein
Flavoproteins are proteins that contain a nucleic acid derivative of riboflavin. These proteins are involved in a wide array of biological processes, including removal of radicals contributing to oxidative stress, photosynthesis, and DNA repair. ...
* Mitochondrion
A mitochondrion () is an organelle found in the cell (biology), cells of most eukaryotes, such as animals, plants and fungi. Mitochondria have a double lipid bilayer, membrane structure and use aerobic respiration to generate adenosine tri ...
* Oxidative phosphorylation
Oxidative phosphorylation(UK , US : or electron transport-linked phosphorylation or terminal oxidation, is the metabolic pathway in which Cell (biology), cells use enzymes to Redox, oxidize nutrients, thereby releasing chemical energy in order ...
* P-ATPase
* Proton pump
A proton pump is an integral membrane protein pump that builds up a proton gradient across a biological membrane. Proton pumps catalyze the following reaction:
: n one side of a biological membrane/sub> + energy n the other side of the m ...
* Rotating locomotion in living systems
Several organisms are capable of rolling locomotion. However, true wheels and propellers—despite their utility in human vehicles—do not play a significant role in the movement of living things (with the exception of the corkscrew-like flag ...
* Transmembrane ATPase
* V-ATPase
Vacuolar-type ATPase (V-ATPase) is a highly conserved evolutionarily ancient enzyme with remarkably diverse functions in eukaryotic organisms. V-ATPases acidify a wide array of intracellular organelles and pump protons across the plasma ...
References
Further reading
* Nick Lane''The Vital Question: Energy, Evolution, and the Origins of Complex Life''
Ww Norton, 2015-07-20, (Link points to Figure 10 showing model of ATP synthase)
External links
* Boris A. Feniouk"ATP synthase — a splendid molecular machine"
* Well illustrate
by Antony Crofts of the
University of Illinois at Urbana–Champaign
The University of Illinois Urbana-Champaign (UIUC, U of I, Illinois, or University of Illinois) is a public land-grant research university in the Champaign–Urbana metropolitan area, Illinois, United States. Established in 1867, it is the f ...
.
Proton and Sodium translocating F-type, V-type and A-type ATPases in OPM database
to Paul D. Boyer and John E. Walker for the enzymatic mechanism of synthesis of ATP; and to Jens C. Skou, for discovery of an ion-transporting enzyme, , -ATPase.
– ATP synthesis animation * David Goodsell
"ATP Synthase- Molecule of the Month"
{{Portal bar, Biology, border=no Enzymes Cellular respiration Photosynthesis EC 3.6.3 Integral membrane proteins Protein complexes