|
Transmembrane Topology
Topology of a transmembrane protein refers to locations of N- and C-termini of membrane-spanning polypeptide chain with respect to the inner or outer sides of the biological membrane occupied by the protein. Several databases provide experimentally determined topologies of membrane proteins. They include Uniprot, TOPDB, OPM, and ExTopoDB. There is also a database of domains located conservatively on a certain side of membranes, TOPDOM. Several computational methods were developed, with a limited success, for predicting transmembrane alpha-helices and their topology. Pioneer methods utilized the fact that membrane-spanning regions contain more hydrophobic residues than other parts of the protein, however applying different hydrophobic scales altered the prediction results. Later, several statistical methods were developed to improve the topography prediction and a special alignment method was introduced. According to the positive-inside rule, cytosolic loops near the lipid bilaye ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transmembrane Protein
A transmembrane protein (TP) is a type of integral membrane protein that spans the entirety of the cell membrane. Many transmembrane proteins function as gateways to permit the transport of specific substances across the membrane. They frequently undergo significant conformational changes to move a substance through the membrane. They are usually highly hydrophobic and aggregate and precipitate in water. They require detergents or nonpolar solvents for extraction, although some of them ( beta-barrels) can be also extracted using denaturing agents. The peptide sequence that spans the membrane, or the transmembrane segment, is largely hydrophobic and can be visualized using the hydropathy plot. Depending on the number of transmembrane segments, transmembrane proteins can be classified as single-span (or bitopic) or multi-span (polytopic). Some other integral membrane proteins are called monotopic, meaning that they are also permanently attached to the membrane, but do not ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Biological Membrane
A biological membrane, biomembrane or cell membrane is a selectively permeable membrane that separates the interior of a cell from the external environment or creates intracellular compartments by serving as a boundary between one part of the cell and another. Biological membranes, in the form of eukaryotic cell membranes, consist of a phospholipid bilayer with embedded, integral and peripheral proteins used in communication and transportation of chemicals and ions. The bulk of lipids in a cell membrane provides a fluid matrix for proteins to rotate and laterally diffuse for physiological functioning. Proteins are adapted to high membrane fluidity environment of the lipid bilayer with the presence of an annular lipid shell, consisting of lipid molecules bound tightly to the surface of integral membrane proteins. The cell membranes are different from the isolating tissues formed by layers of cells, such as mucous membranes, basement membranes, and serous membranes. C ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
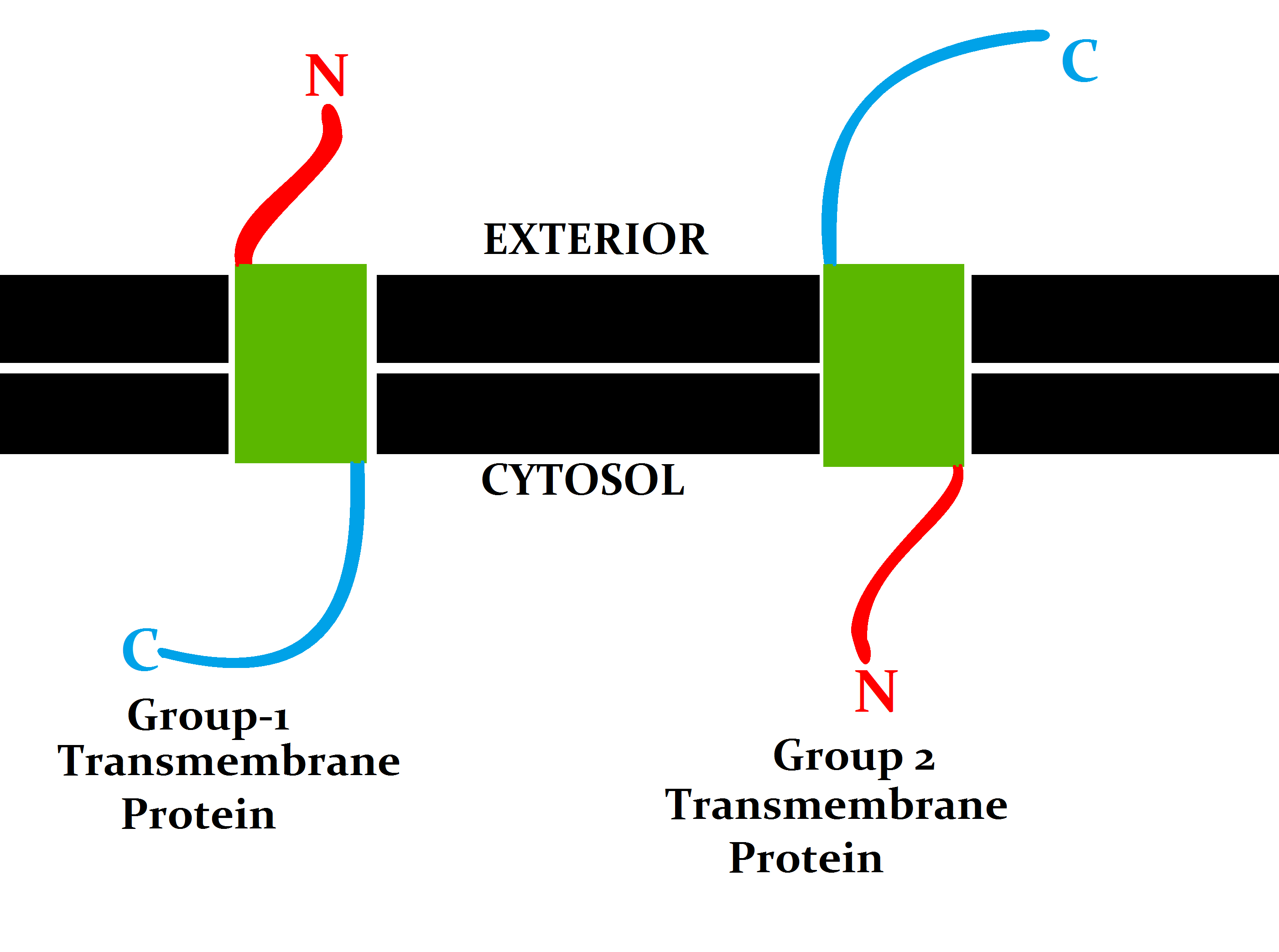
Group 1 And 2 Transmembrane Protein
A group is a number of persons or things that are located, gathered, or classed together. Groups of people * Cultural group, a group whose members share the same cultural identity * Ethnic group, a group whose members share the same ethnic identity * Religious group (other), a group whose members share the same religious identity * Social group, a group whose members share the same social identity * Tribal group, a group whose members share the same tribal identity * Organization, an entity that has a collective goal and is linked to an external environment * Peer group, an entity of three or more people with similar age, ability, experience, and interest Social science * In-group and out-group * Primary, secondary, and reference groups * Social group * Collectives Science and technology Mathematics * Group (mathematics), a set together with a binary operation satisfying certain algebraic conditions Chemistry * Functional group, a group of atoms which provide ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Uniprot
UniProt is a freely accessible database of protein sequence and functional information, many entries being derived from genome sequencing projects. It contains a large amount of information about the biological function of proteins derived from the research literature. It is maintained by the UniProt consortium, which consists of several European bioinformatics organisations and a foundation from Washington, DC, United States. The UniProt consortium The UniProt consortium comprises the European Bioinformatics Institute (EBI), the Swiss Institute of Bioinformatics (SIB), and the Protein Information Resource (PIR). EBI, located at the Wellcome Trust Genome Campus in Hinxton, UK, hosts a large resource of bioinformatics databases and services. SIB, located in Geneva, Switzerland, maintains the ExPASy (Expert Protein Analysis System) servers that are a central resource for proteomics tools and databases. PIR, hosted by the National Biomedical Research Foundation (NBRF) at the Geo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Orientations Of Proteins In Membranes Database
Orientations of Proteins in Membranes (OPM) database provides spatial positions of membrane protein structures with respect to the lipid bilayer. Positions of the proteins are calculated using an implicit solvation model of the lipid bilayer. The results of calculations were verified against experimental studies of spatial arrangement of transmembrane and peripheral proteins in membranes. Proteins structures are taken from the Protein Data Bank. OPM also provides structural classification of membrane-associated proteins into families and superfamilies, membrane topology, quaternary structure of proteins in membrane-bound state, and the type of a destination membrane for each protein. The coordinate files with calculated membrane boundaries are downloadable. The site allows visualization of protein structures with membrane boundary planes through Jmol. The database was widely used in experimental and theoretical studies of membrane-associated proteins. However, structures o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
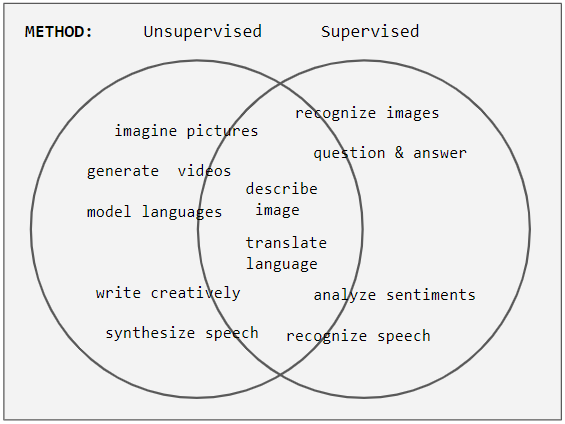
Supervised Learning
Supervised learning (SL) is a machine learning paradigm for problems where the available data consists of labelled examples, meaning that each data point contains features (covariates) and an associated label. The goal of supervised learning algorithms is learning a function that maps feature vectors (inputs) to labels (output), based on example input-output pairs. It infers a function from ' consisting of a set of ''training examples''. In supervised learning, each example is a ''pair'' consisting of an input object (typically a vector) and a desired output value (also called the ''supervisory signal''). A supervised learning algorithm analyzes the training data and produces an inferred function, which can be used for mapping new examples. An optimal scenario will allow for the algorithm to correctly determine the class labels for unseen instances. This requires the learning algorithm to generalize from the training data to unseen situations in a "reasonable" way (see inductive ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Unsupervised Learning
Unsupervised learning is a type of algorithm that learns patterns from untagged data. The hope is that through mimicry, which is an important mode of learning in people, the machine is forced to build a concise representation of its world and then generate imaginative content from it. In contrast to supervised learning where data is tagged by an expert, e.g. tagged as a "ball" or "fish", unsupervised methods exhibit self-organization that captures patterns as probability densities or a combination of neural feature preferences encoded in the machine's weights and activations. The other levels in the supervision spectrum are reinforcement learning where the machine is given only a numerical performance score as guidance, and semi-supervised learning where a small portion of the data is tagged. Neural networks Tasks vs. methods Neural network tasks are often categorized as discriminative (recognition) or generative (imagination). Often but not always, discriminative tas ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Signal Peptide
A signal peptide (sometimes referred to as signal sequence, targeting signal, localization signal, localization sequence, transit peptide, leader sequence or leader peptide) is a short peptide (usually 16-30 amino acids long) present at the N-terminus (or occasionally nonclassically at the C-terminus or internally) of most newly synthesized proteins that are destined toward the secretory pathway. These proteins include those that reside either inside certain organelles (the endoplasmic reticulum, Golgi or endosomes), secreted from the cell, or inserted into most cellular membranes. Although most type I membrane-bound proteins have signal peptides, the majority of type II and multi-spanning membrane-bound proteins are targeted to the secretory pathway by their first transmembrane domain, which biochemically resembles a signal sequence except that it is not cleaved. They are a kind of target peptide. Function (translocation) Signal peptides function to prompt a cell to tra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Topology
Protein topology is a property of protein molecule that does not change under deformation (without cutting or breaking a bond). Frameworks Two main topology frameworks have been developed and applied to protein molecules. Knot Theory Knot theory which categorises chain entanglements. The usage of knot theory is limited to a small percentage of proteins as most of them are unknot. Circuit topology Circuit topology categorises intra-chain contacts based on their arrangements. Other Uses In biology literature, the term topology is also used to refer to mutual orientation of regular secondary structures, such as alpha-helices and beta strands in protein structure For example, two adjacent interacting alpha-helices or beta-strands can go in the same or in opposite directions. Topology diagrams of dif ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transmembrane Protein
A transmembrane protein (TP) is a type of integral membrane protein that spans the entirety of the cell membrane. Many transmembrane proteins function as gateways to permit the transport of specific substances across the membrane. They frequently undergo significant conformational changes to move a substance through the membrane. They are usually highly hydrophobic and aggregate and precipitate in water. They require detergents or nonpolar solvents for extraction, although some of them ( beta-barrels) can be also extracted using denaturing agents. The peptide sequence that spans the membrane, or the transmembrane segment, is largely hydrophobic and can be visualized using the hydropathy plot. Depending on the number of transmembrane segments, transmembrane proteins can be classified as single-span (or bitopic) or multi-span (polytopic). Some other integral membrane proteins are called monotopic, meaning that they are also permanently attached to the membrane, but do not ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Integral Membrane Protein
An integral, or intrinsic, membrane protein (IMP) is a type of membrane protein that is permanently attached to the biological membrane. All ''transmembrane proteins'' are IMPs, but not all IMPs are transmembrane proteins. IMPs comprise a significant fraction of the proteins encoded in an organism's genome. Proteins that cross the membrane are surrounded by annular lipids, which are defined as lipids that are in direct contact with a membrane protein. Such proteins can only be separated from the membranes by using detergents, nonpolar solvents, or sometimes denaturing agents. Structure Three-dimensional structures of ~160 different integral membrane proteins have been determined at atomic resolution by X-ray crystallography or nuclear magnetic resonance spectroscopy. They are challenging subjects for study owing to the difficulties associated with extraction and crystallization. In addition, structures of many water- soluble protein domains of IMPs are available in the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Endomembrane System
The endomembrane system is composed of the different membranes (endomembranes) that are suspended in the cytoplasm within a eukaryotic cell. These membranes divide the cell into functional and structural compartments, or organelles. In eukaryotes the organelles of the endomembrane system include: the nuclear membrane, the endoplasmic reticulum, the Golgi apparatus, lysosomes, vesicles, endosomes, and plasma (cell) membrane among others. The system is defined more accurately as the set of membranes that forms a single functional and developmental unit, either being connected directly, or exchanging material through vesicle transport. Importantly, the endomembrane system does not include the membranes of plastids or mitochondria, but might have evolved partially from the actions of the latter (see below). The nuclear membrane contains a lipid bilayer that encompasses the contents of the nucleus. The endoplasmic reticulum (ER) is a synthesis and transport organelle that branches ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |