|
Transglutaminases
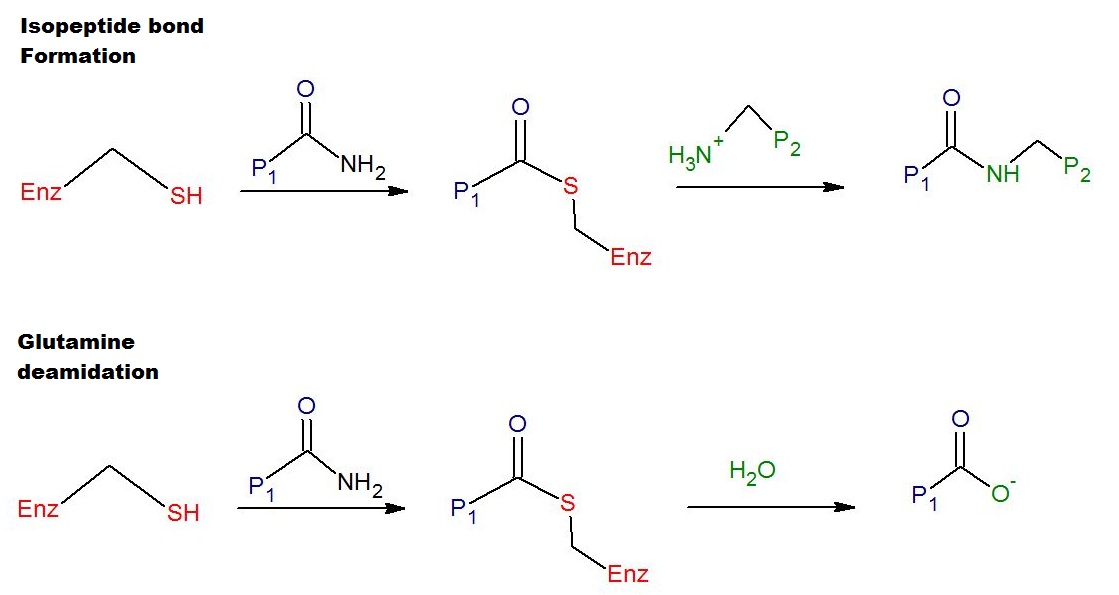
Transglutaminases are enzymes that in nature primarily catalyze the formation of an isopeptide bond between γ-carboxamide groups ( -(C=O)NH2 ) of glutamine residue side chains and the ε-amino groups ( -NH2 ) of lysine residue side chains with subsequent release of ammonia ( NH3 ). Lysine and glutamine residues must be bound to a peptide or a protein so that this cross-linking (between separate molecules) or intramolecular (within the same molecule) reaction can happen. Bonds formed by transglutaminase exhibit high resistance to proteolytic degradation (proteolysis). The reaction is :Glutamine, Gln-(C=O)NH2 + Lysine, NH2-Lys → Gln-(C=O)NH-Lys + NH3 Transglutaminases can also join a primary amine ( RNH2 ) to the side chain carboxyamide group of a protein/peptide bound glutamine residue thus forming an isopeptide bond :Gln-(C=O)NH2 + RNH2 → Gln-(C=O)NHR + NH3 These enzymes can also Deamidation, deamidate glutamine residues to glutamic acid residues ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are catalytic RNA molecules, called ribozymes. Enzymes' specificity comes from their unique three-dimensional structures. Like all catalysts, enzymes increase the reaction ra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Glutamic Acid
Glutamic acid (symbol Glu or E; the ionic form is known as glutamate) is an α-amino acid that is used by almost all living beings in the biosynthesis of proteins. It is a non-essential nutrient for humans, meaning that the human body can synthesize enough for its use. It is also the most abundant excitatory neurotransmitter in the vertebrate nervous system. It serves as the precursor for the synthesis of the inhibitory gamma-aminobutyric acid (GABA) in GABA-ergic neurons. Its molecular formula is . Glutamic acid exists in three optically isomeric forms; the dextrorotatory -form is usually obtained by hydrolysis of gluten or from the waste waters of beet-sugar manufacture or by fermentation.Webster's Third New International Dictionary of the English Language Unabridged, Third Edition, 1971. Its molecular structure could be idealized as HOOC−CH()−()2−COOH, with two carboxyl groups −COOH and one amino group −. However, in the solid state and mildly acidic water solutio ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Catalytic Triad
A catalytic triad is a set of three coordinated amino acids that can be found in the active site of some enzymes. Catalytic triads are most commonly found in hydrolase and transferase enzymes (e.g. proteases, amidases, esterases, acylases, lipases and β-lactamases). An acid- base-nucleophile triad is a common motif for generating a nucleophilic residue for covalent catalysis. The residues form a charge-relay network to polarise and activate the nucleophile, which attacks the substrate, forming a covalent intermediate which is then hydrolysed to release the product and regenerate free enzyme. The nucleophile is most commonly a serine or cysteine amino acid, but occasionally threonine or even selenocysteine. The 3D structure of the enzyme brings together the triad residues in a precise orientation, even though they may be far apart in the sequence (primary structure). As well as divergent evolution of function (and even the triad's nucleophile), catalytic triads show some ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Papain-like Protease
Papain-like proteases (or papain-like (cysteine) peptidases; abbreviated PLP or PLCP) are a large protein family of cysteine protease enzymes that share structural and enzymatic properties with the group's namesake member, papain. They are found in all domains of life. In animals, the group is often known as cysteine cathepsins or, in older literature, lysosomal peptidases. In the MEROPS protease enzyme classification system, papain-like proteases form Clan CA. Papain-like proteases share a common catalytic dyad active site featuring a cysteine amino acid residue that acts as a nucleophile. The human genome encodes eleven cysteine cathepsins which have a broad range of physiological functions. In some parasites papain-like proteases have roles in host invasion, such as cruzipain from ''Trypanosoma cruzi''. In plants, they are involved in host defense and in development. Studies of papain-like proteases from prokaryotes have lagged their eukaryotic counterparts. In cellular organ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Immunoglobulin
An antibody (Ab), also known as an immunoglobulin (Ig), is a large, Y-shaped protein used by the immune system to identify and neutralize foreign objects such as pathogenic bacteria and viruses. The antibody recognizes a unique molecule of the pathogen, called an antigen. Each tip of the "Y" of an antibody contains a paratope (analogous to a lock) that is specific for one particular epitope (analogous to a key) on an antigen, allowing these two structures to bind together with precision. Using this binding mechanism, an antibody can ''tag'' a microbe or an infected cell for attack by other parts of the immune system, or can neutralize it directly (for example, by blocking a part of a virus that is essential for its invasion). To allow the immune system to recognize millions of different antigens, the antigen-binding sites at both tips of the antibody come in an equally wide variety. In contrast, the remainder of the antibody is relatively constant. It only occurs in a few vari ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transamidation
Transamidation is a chemical reaction in which an amide reacts with an amine to generate a new amide: :RC(O)NR'2 + HNR"2 → RC(O)NR"2 + HNR'2 The reaction is typically very slow, but it can be accelerated with Lewis acid and organometallic catalysts. Primary amides (RC(O)NH2) are more amenable to this reaction. Ureas In contrast to the reluctance of amides as substrates, urea is more susceptible to this exchange process. Transamidation is practiced, sometimes even on an industrial scale, to prepare a variety of N-substituted ureas: :(H2N)2CO + R2NH → (R2N)(H2N)CO + NH3 :(R2N)(H2N)CO + R2NH → (R2N)2CO + NH3 Methylurea, precursor to theobromine, is produced from methylamine and urea. Phenylurea is produced similarly but from anilinium chloride:{{cite journal , authors= , title=Arylureas II. Urea Method p-Ethoxyphenylurea , journal=Org. Synth. , year=1951 , volume=31 , page=11 , doi=10.15227/orgsyn.031.0011 :(H2N)2CO + 6H5NH3l → (C6H5(H)N)(H2N) ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transamidation And Deamidation Mechanisms Of Tissue Transglutaminase
Transamidation is a chemical reaction in which an amide reacts with an amine to generate a new amide: :RC(O)NR'2 + HNR"2 → RC(O)NR"2 + HNR'2 The reaction is typically very slow, but it can be accelerated with Lewis acid and organometallic catalysts. Primary amides (RC(O)NH2) are more amenable to this reaction. Ureas In contrast to the reluctance of amides as substrates, urea is more susceptible to this exchange process. Transamidation is practiced, sometimes even on an industrial scale, to prepare a variety of N-substituted ureas: :(H2N)2CO + R2NH → (R2N)(H2N)CO + NH3 :(R2N)(H2N)CO + R2NH → (R2N)2CO + NH3 Methylurea, precursor to theobromine, is produced from methylamine and urea. Phenylurea is produced similarly but from anilinium chloride:{{cite journal , authors= , title=Arylureas II. Urea Method p-Ethoxyphenylurea , journal=Org. Synth. , year=1951 , volume=31 , page=11 , doi=10.15227/orgsyn.031.0011 :(H2N)2CO + 6H5NH3l → (C6H5(H)N)(H2N) ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |