|
TRNASer (uridine44-2'-O)-methyltransferase
TRNASer (uridine44-2'-O)-methyltransferase (, ''TRM44'') is an enzyme with systematic name ''S-adenosyl-L-methionine:tRNASer (uridine44-2'-O)-methyltransferase''. This enzyme catalyses the following chemical reaction : S-adenosyl-L-methionine + uridine44 in tRNASer \rightleftharpoons S-adenosyl-L-homocysteine + 2'-O-methyluridine44 in tRNASer The 2'-O-methylation of uridine44 contributes to stability of tRNASer(CGA). References External links * {{Portal bar, Biology, border=no EC 2.1.1 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are catalytic RNA molecules, called ribozymes. Enzymes' specificity comes from their unique three-dimensional structures. Like all catalysts, enzymes increase the reaction ra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of Enzymes
This article lists enzymes by their classification in the International Union of Biochemistry and Molecular Biology's Enzyme Commission (EC) numbering system. * List of EC numbers (EC 5) * List of EC numbers (EC 6) :Oxidoreductases (EC 1) (Oxidoreductase) *Dehydrogenase * Luciferase *DMSO reductase :EC 1.1 (act on the CH-OH group of donors) * :EC 1.1.1 (with NAD+ or NADP+ as acceptor) ** Alcohol dehydrogenase (NAD) ** Alcohol dehydrogenase (NADP) **Homoserine dehydrogenase ** Aminopropanol oxidoreductase **Diacetyl reductase **Glycerol dehydrogenase **Propanediol-phosphate dehydrogenase ** glycerol-3-phosphate dehydrogenase (NAD+) ** D-xylulose reductase **L-xylulose reductase **Lactate dehydrogenase **Malate dehydrogenase **Isocitrate dehydrogenase ** HMG-CoA reductase * :EC 1.1.2 (with a cytochrome as acceptor) * :EC 1.1.3 (with oxygen as acceptor) **Glucose oxidase **L-gulonolactone oxidase **Thiamine oxidase **Xanthine oxidase * :EC 1.1.4 (with a disul ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Catalysis
Catalysis () is the process of increasing the rate of a chemical reaction by adding a substance known as a catalyst (). Catalysts are not consumed in the reaction and remain unchanged after it. If the reaction is rapid and the catalyst recycles quickly, very small amounts of catalyst often suffice; mixing, surface area, and temperature are important factors in reaction rate. Catalysts generally react with one or more reactants to form intermediates that subsequently give the final reaction product, in the process of regenerating the catalyst. Catalysis may be classified as either homogeneous, whose components are dispersed in the same phase (usually gaseous or liquid) as the reactant, or heterogeneous, whose components are not in the same phase. Enzymes and other biocatalysts are often considered as a third category. Catalysis is ubiquitous in chemical industry of all kinds. Estimates are that 90% of all commercially produced chemical products involve catalysts at some s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chemical Reaction
A chemical reaction is a process that leads to the IUPAC nomenclature for organic transformations, chemical transformation of one set of chemical substances to another. Classically, chemical reactions encompass changes that only involve the positions of electrons in the forming and breaking of chemical bonds between atoms, with no change to the Atomic nucleus, nuclei (no change to the elements present), and can often be described by a chemical equation. Nuclear chemistry is a sub-discipline of chemistry that involves the chemical reactions of unstable and radioactive Chemical element, elements where both electronic and nuclear changes can occur. The substance (or substances) initially involved in a chemical reaction are called reagent, reactants or reagents. Chemical reactions are usually characterized by a chemical change, and they yield one or more Product (chemistry), products, which usually have properties different from the reactants. Reactions often consist of a sequence o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
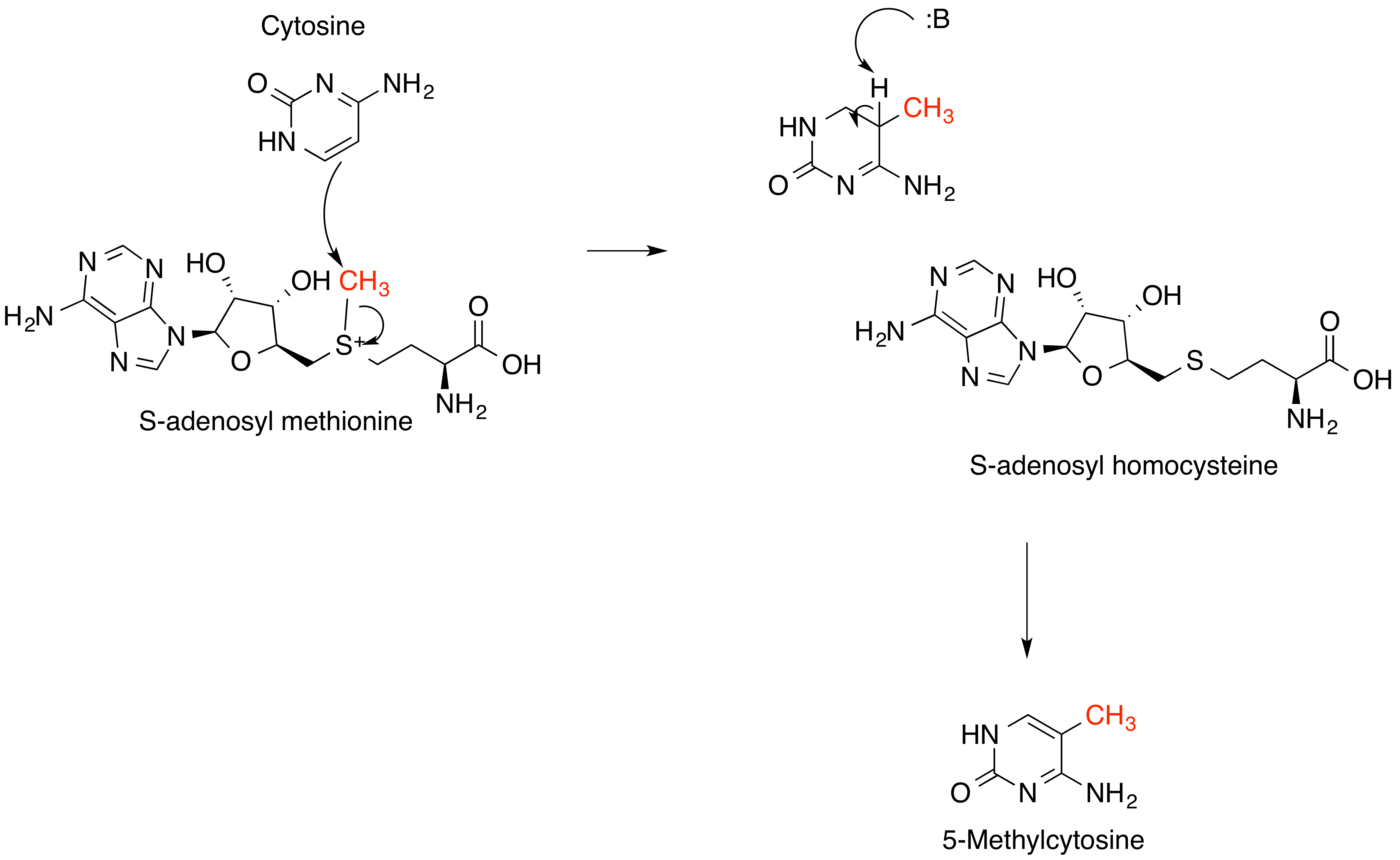
S-adenosyl-L-methionine
''S''-Adenosyl methionine (SAM), also known under the commercial names of SAMe, SAM-e, or AdoMet, is a common cosubstrate involved in methyl group transfers, transsulfuration, and aminopropylation. Although these anabolic reactions occur throughout the body, most SAM is produced and consumed in the liver. More than 40 methyl transfers from SAM are known, to various substrates such as nucleic acids, proteins, lipids and secondary metabolites. It is made from adenosine triphosphate (ATP) and methionine by methionine adenosyltransferase. SAM was first discovered by Giulio Cantoni in 1952. In bacteria, SAM is bound by the SAM riboswitch, which regulates genes involved in methionine or cysteine biosynthesis. In eukaryotic cells, SAM serves as a regulator of a variety of processes including DNA, tRNA, and rRNA methylation; immune response; amino acid metabolism; transsulfuration; and more. In plants, SAM is crucial to the biosynthesis of ethylene, an important plant hormone and sign ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Uridine
Uridine (symbol U or Urd) is a glycosylated pyrimidine analog containing uracil attached to a ribose ring (or more specifically, a ribofuranose) via a β-N1-glycosidic bond. The analog is one of the five standard nucleosides which make up nucleic acids, the others being adenosine, thymidine, cytidine and guanosine. The five nucleosides are commonly abbreviated to their symbols, U, A, dT, C, and G, respectively. However, thymidine is more commonly written as 'dT' ('d' represents 'deoxy') as it contains a 2'-deoxyribofuranose moiety rather than the ribofuranose ring found in uridine. This is because thymidine is found in deoxyribonucleic acid (DNA) and usually not in ribonucleic acid (RNA). Conversely, uridine is found in RNA and not DNA. The remaining three nucleosides may be found in both RNA and DNA. In RNA, they would be represented as A, C and G whereas in DNA they would be represented as dA, dC and dG. Biosynthesis Uridine is widely produced in nature as uridine monophosp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Serine—tRNA Ligase
In enzymology, a serine—tRNA ligase () is an enzyme that catalyzes the chemical reaction :ATP + L-serine + tRNASer \rightleftharpoons AMP + diphosphate + L-seryl-tRNASer The 3 substrates of this enzyme are ATP, L-serine, and tRNA(Ser), whereas its 3 products are AMP, diphosphate, and L-seryl-tRNA(Ser). This enzyme belongs to the family of ligases, to be specific those forming carbon-oxygen bonds in aminoacyl-tRNA and related compounds. The systematic name of this enzyme class is L-serine:tRNASer ligase (AMP-forming). Other names in common use include seryl-tRNA synthetase, SerRS, seryl-transfer ribonucleate synthetase, seryl-transfer RNA synthetase, seryl-transfer ribonucleic acid synthetase, and serine translase. This enzyme participates in glycine, serine and threonine metabolism and aminoacyl-trna biosynthesis Aminoacyl-tRNA (also aa-tRNA or charged tRNA) is tRNA to which its cognate amino acid is chemically bonded (charged). The aa-tRNA, along with particular ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |