|
Substrate Specificity
Chemical specificity is the ability of binding site of a macromolecule (such as a protein) to bind specific ligands. The fewer ligands a protein can bind, the greater its specificity. Specificity describes the strength of binding between a given protein and ligand. This relationship can be described by a dissociation constant, which characterizes the balance between bound and unbound states for the protein-ligand system. In the context of a single enzyme and a pair of binding molecules, the two ligands can be compared as stronger or weaker ligands (for the enzyme) on the basis of their dissociation constants. (A lower value corresponds to a stronger binding.) Specificity for a set of ligands is unrelated to the ability of an enzyme to catalyze a given reaction, with the ligand as a substrate. If a given enzyme has a high chemical specificity, this means that the set of ligands to which it binds is limited, such that neither binding events nor catalysis can occur at an appreci ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Binding Site
In biochemistry and molecular biology, a binding site is a region on a macromolecule such as a protein that binds to another molecule with specificity. The binding partner of the macromolecule is often referred to as a ligand. Ligands may include other proteins (resulting in a protein-protein interaction), enzyme substrates, second messengers, hormones, or allosteric modulators. The binding event is often, but not always, accompanied by a conformational change that alters the protein's function. Binding to protein binding sites is most often reversible (transient and non-covalent), but can also be covalent reversible or irreversible. Function Binding of a ligand to a binding site on protein often triggers a change in conformation in the protein and results in altered cellular function. Hence binding site on protein are critical parts of signal transduction pathways. Types of ligands include neurotransmitters, toxins, neuropeptides, and steroid hormones. Binding sites incur ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Electrostatic Interactions
Electrostatics is a branch of physics that studies electric charges at Rest (physics), rest (static electricity). Since classical antiquity, classical times, it has been known that some materials, such as amber, attract lightweight particles after triboelectric effect, rubbing. The Greek language, Greek word for amber, (), was thus the source of the word 'electricity'. Electrostatic phenomena arise from the forces that electric charges exert on each other. Such forces are described by Coulomb's law. Even though electrostatically induced forces seem to be rather weak, some electrostatic forces are relatively large. The force between an electron and a proton, which together make up a hydrogen atom, is about 36 order of magnitude, orders of magnitude stronger than the gravitational force acting between them. There are many examples of electrostatic phenomena, from those as simple as the attraction of plastic wrap to one's hand after it is removed from a package, to the apparently ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Substrate (chemistry)
In chemistry, the term substrate is highly context-dependent. Broadly speaking, it can refer either to a chemical species being observed in a chemical reaction, or to a surface on which other chemical reactions or microscopy are performed. In the former sense, a reagent is added to the ''substrate'' to generate a product through a chemical reaction. The term is used in a similar sense in synthetic and organic chemistry, where the substrate is the chemical of interest that is being modified. In biochemistry, an enzyme substrate is the material upon which an enzyme acts. When referring to Le Chatelier's principle, the substrate is the reagent whose concentration is changed. ;Spontaneous reaction : :*Where S is substrate and P is product. ;Catalysed reaction : :*Where S is substrate, P is product and C is catalyst. In the latter sense, it may refer to a surface on which other chemical reactions are performed or play a supporting role in a variety of spectroscopic and micro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Antibody
An antibody (Ab), also known as an immunoglobulin (Ig), is a large, Y-shaped protein used by the immune system to identify and neutralize foreign objects such as pathogenic bacteria and viruses. The antibody recognizes a unique molecule of the pathogen, called an antigen. Each tip of the "Y" of an antibody contains a paratope (analogous to a lock) that is specific for one particular epitope (analogous to a key) on an antigen, allowing these two structures to bind together with precision. Using this binding mechanism, an antibody can ''tag'' a microbe or an infected cell for attack by other parts of the immune system, or can neutralize it directly (for example, by blocking a part of a virus that is essential for its invasion). To allow the immune system to recognize millions of different antigens, the antigen-binding sites at both tips of the antibody come in an equally wide variety. In contrast, the remainder of the antibody is relatively constant. It only occurs in a few vari ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ENZYME STRUCTURE AND FUNCTION
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are catalytic RNA molecules, called ribozymes. Enzymes' specificity comes from their unique three-dimensional structures. Like all catalysts, enzymes increase the reaction ra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Specificity Constant
In the field of biochemistry, the specificity constant (also called kinetic efficiency or k_/K_), is a measure of how efficiently an enzyme converts substrates into products. A comparison of specificity constants can also be used as a measure of the preference of an enzyme for different substrates (i.e., substrate specificity). The higher the specificity constant, the more the enzyme "prefers" that substrate. The following equation, known as the Michaelis–Menten model, is used to describe the kinetics of enzymes: : + S _fk_r] ES -> _ + P where E, S, ES, and P represent enzyme, substrate, enzyme–substrate complex, and product, respectively. The symbols k_f, k_r, and k_\mathrm denote the rate constants for the "forward" binding and "reverse" unbinding of substrate, and for the "catalytic" conversion of substrate into product, respectively. The Michaelis constant in turn is defined as follows: : K_ = \frac The Michaelis constant is equal to the substrate concentration at ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Amylase
An amylase () is an enzyme that catalyses the hydrolysis of starch (Latin ') into sugars. Amylase is present in the saliva of humans and some other mammals, where it begins the chemical process of digestion. Foods that contain large amounts of starch but little sugar, such as rice and potatoes, may acquire a slightly sweet taste as they are chewed because amylase degrades some of their starch into sugar. The pancreas and salivary gland make amylase ( alpha amylase) to hydrolyse dietary starch into disaccharides and trisaccharides which are converted by other enzymes to glucose to supply the body with energy. Plants and some bacteria also produce amylase. Specific amylase proteins are designated by different Greek letters. All amylases are glycoside hydrolases and act on α-1,4-glycosidic bonds. Classification α-Amylase The α-amylases () (CAS 9014-71-5) (alternative names: 1,4-α-D-glucan glucanohydrolase; glycogenase) are calcium metalloenzymes. By acting at random loc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
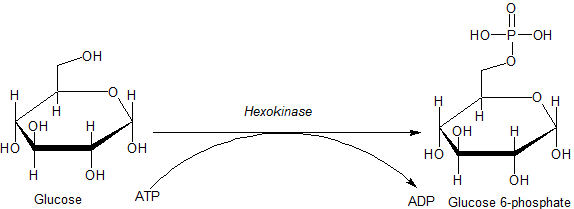
Hexokinase
A hexokinase is an enzyme that phosphorylates hexoses (six-carbon sugars), forming hexose phosphate. In most organisms, glucose is the most important substrate for hexokinases, and glucose-6-phosphate is the most important product. Hexokinase possesses the ability to transfer an inorganic phosphate group from ATP to a substrate. Hexokinases should not be confused with glucokinase, which is a specific isoform of hexokinase. All hexokinases are capable of phosphorylating several hexoses but glucokinase acts with a 50-fold lower substrate affinity and its main hexose substrate is glucose. Variation Genes that encode hexokinase have been discovered in every domain of life, and exist among a variety of species that range from bacteria, yeast, and plants to humans and other vertebrates. They are categorized as ''actin fold'' proteins, sharing a common ATP binding site core that is surrounded by more variable sequences which determine substrate affinities and other propertie ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
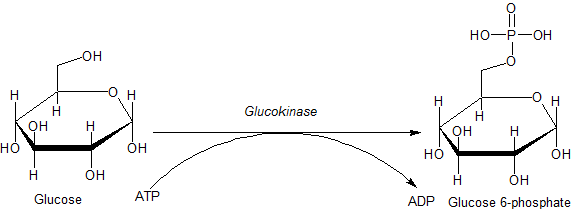
Glucokinase
Glucokinase () is an enzyme that facilitates phosphorylation of glucose to glucose-6-phosphate. Glucokinase occurs in cells in the liver and pancreas of humans and most other vertebrates. In each of these organs it plays an important role in the regulation of carbohydrate metabolism by acting as a glucose sensor, triggering shifts in metabolism or cell function in response to rising or falling levels of glucose, such as occur after a meal or when fasting. Mutations of the gene for this enzyme can cause unusual forms of diabetes or hypoglycemia. Glucokinase (GK) is a hexokinase isozyme, related homologously to at least three other hexokinases. All of the hexokinases can mediate phosphorylation of glucose to glucose-6-phosphate (G6P), which is the first step of both glycogen synthesis and glycolysis. However, glucokinase is coded by a separate gene and its distinctive kinetic properties allow it to serve a different set of functions. Glucokinase has a lower affinity for g ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hydrophobic Interactions
The hydrophobic effect is the observed tendency of nonpolar substances to aggregate in an aqueous solution and exclude water#Properties, water molecules. The word hydrophobic literally means "water-fearing", and it describes the Segregation in materials, segregation of water and nonpolar substances, which maximizes hydrogen bonding between molecules of water and minimizes the area of contact between water and nonpolar molecules. In terms of thermodynamics, the hydrophobic effect is the free energy change of water surrounding a solute. A positive free energy change of the surrounding solvent indicates hydrophobicity, whereas a negative free energy change implies hydrophilicity. The hydrophobic effect is responsible for the separation of a mixture of oil and water into its two components. It is also responsible for effects related to biology, including: cell membrane and vesicle formation, protein folding, insertion of membrane proteins into the nonpolar lipid environment and prot ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |