|
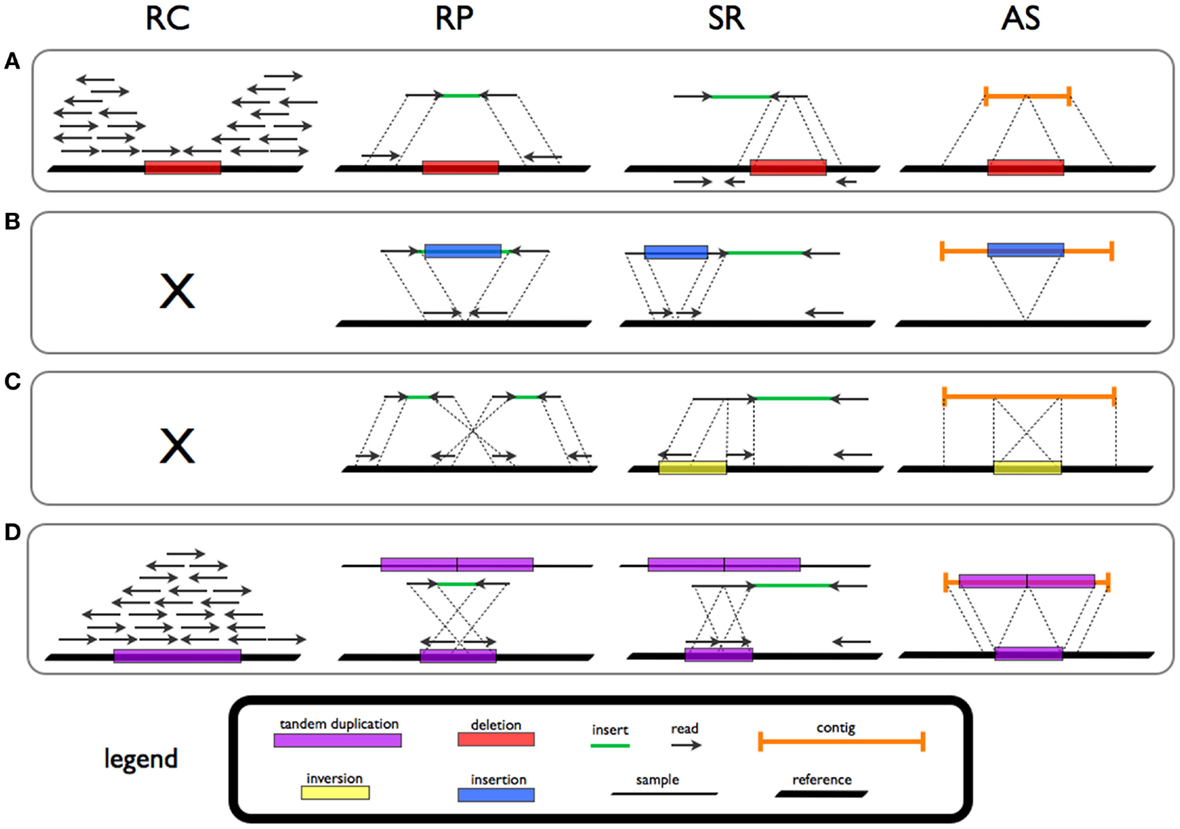
Structural Variations
Genomic structural variation is the variation in structure of an organism's chromosome. It consists of many kinds of variation in the genome of one species, and usually includes microscopic and submicroscopic types, such as deletions, duplications, copy-number variants, insertions, inversions and translocations. Originally, a structure variation affects a sequence length about 1kb to 3Mb, which is larger than SNPs and smaller than chromosome abnormality (though the definitions have some overlap). However, the operational range of structural variants has widened to include events > 50bp. The definition of structural variation does not imply anything about frequency or phenotypical effects. Many structural variants are associated with genetic diseases, however many are not. Recent research about SVs indicates that SVs are more difficult to detect than SNPs. Approximately 13% of the human genome is defined as structurally variant in the normal population, and there are at least 2 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
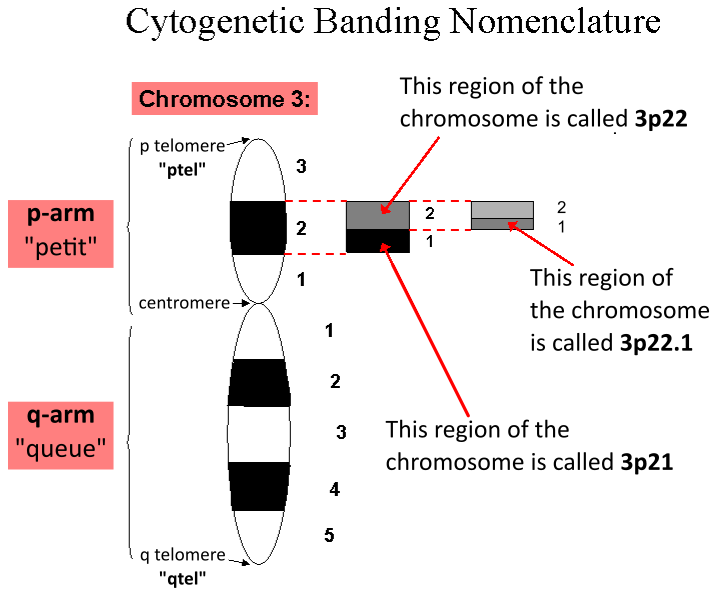
Chromosome
A chromosome is a long DNA molecule with part or all of the genetic material of an organism. In most chromosomes the very long thin DNA fibers are coated with packaging proteins; in eukaryotic cells the most important of these proteins are the histones. These proteins, aided by chaperone proteins, bind to and condense the DNA molecule to maintain its integrity. These chromosomes display a complex three-dimensional structure, which plays a significant role in transcriptional regulation. Chromosomes are normally visible under a light microscope only during the metaphase of cell division (where all chromosomes are aligned in the center of the cell in their condensed form). Before this happens, each chromosome is duplicated ( S phase), and both copies are joined by a centromere, resulting either in an X-shaped structure (pictured above), if the centromere is located equatorially, or a two-arm structure, if the centromere is located distally. The joined copies are now called si ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Locus (genetics)
In genetics, a locus (plural loci) is a specific, fixed position on a chromosome where a particular gene or genetic marker is located. Each chromosome carries many genes, with each gene occupying a different position or locus; in humans, the total number of protein-coding genes in a complete haploid set of 23 chromosomes is estimated at 19,000–20,000. Genes may possess multiple variants known as alleles, and an allele may also be said to reside at a particular locus. Diploid and polyploid cells whose chromosomes have the same allele at a given locus are called homozygous with respect to that locus, while those that have different alleles at a given locus are called heterozygous. The ordered list of loci known for a particular genome is called a gene map. Gene mapping is the process of determining the specific locus or loci responsible for producing a particular phenotype or biological trait. Association mapping, also known as "linkage disequilibrium mapping", is a method of ma ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Unequal Crossing Over
Unequal crossing over is a type of gene duplication or deletion event that deletes a sequence in one strand and replaces it with a duplication from its sister chromatid in mitosis or from its homologous chromosome during meiosis. It is a type of chromosomal crossover between homologous sequences that are not paired precisely. Normally genes are responsible for occurrence of crossing over. It exchanges sequences of different links between chromosomes. Along with gene conversion, it is believed to be the main driver for the generation of gene duplications and is a source of mutation in the genome. Mechanisms During meiosis, the duplicated chromosomes (chromatids) in eukaryotic organisms are attached to each other in the centromere region and are thus paired. The maternal and paternal chromosomes then align alongside each other. During this time, recombination can take place via crossing over of sections of the paternal and maternal chromatids and leads to reciprocal recombination ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Single Nucleotide Polymorphism
In genetics, a single-nucleotide polymorphism (SNP ; plural SNPs ) is a germline substitution of a single nucleotide at a specific position in the genome. Although certain definitions require the substitution to be present in a sufficiently large fraction of the population (e.g. 1% or more), many publications do not apply such a frequency threshold. For example, at a specific base position in the human genome, the G nucleotide may appear in most individuals, but in a minority of individuals, the position is occupied by an A. This means that there is a SNP at this specific position, and the two possible nucleotide variations – G or A – are said to be the alleles for this specific position. SNPs pinpoint differences in our susceptibility to a wide range of diseases, for example age-related macular degeneration (a common SNP in the CFH gene is associated with increased risk of the disease) or nonalcoholic fatty liver disease (a SNP in the PNPLA3 gene is associated with incr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene Duplication
Gene duplication (or chromosomal duplication or gene amplification) is a major mechanism through which new genetic material is generated during molecular evolution. It can be defined as any duplication of a region of DNA that contains a gene. Gene duplications can arise as products of several types of errors in DNA replication and repair machinery as well as through fortuitous capture by selfish genetic elements. Common sources of gene duplications include ectopic recombination, retrotransposition event, aneuploidy, polyploidy, and replication slippage. Mechanisms of duplication Ectopic recombination Duplications arise from an event termed unequal crossing-over that occurs during meiosis between misaligned homologous chromosomes. The chance of it happening is a function of the degree of sharing of repetitive elements between two chromosomes. The products of this recombination are a duplication at the site of the exchange and a reciprocal deletion. Ectopic recombination is ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
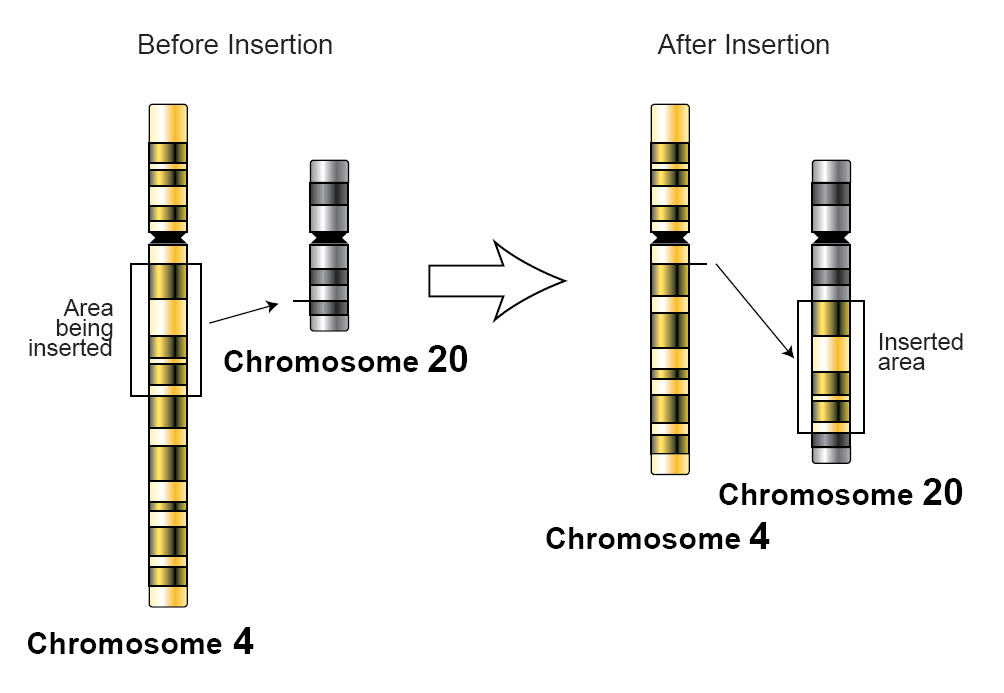
Insertion (genetics)
In genetics, an insertion (also called an insertion mutation) is the addition of one or more nucleotide base pairs into a DNA sequence. This can often happen in microsatellite regions due to the DNA polymerase slipping. Insertions can be anywhere in size from one base pair incorrectly inserted into a DNA sequence to a section of one chromosome inserted into another. The mechanism of the smallest single base insertion mutations is believed to be through base-pair separation between the template and primer strands followed by non-neighbor base stacking, which can occur locally within the DNA polymerase active site. On a chromosome level, an ''insertion'' refers to the insertion of a larger sequence into a chromosome. This can happen due to unequal crossover during meiosis. N region addition is the addition of non-coded nucleotides during recombination by terminal deoxynucleotidyl transferase. P nucleotide insertion is the insertion of palindromic sequences encoded by the ends ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Indel
Indel is a molecular biology term for an insertion or deletion of bases in the genome of an organism. It is classified among small genetic variations, measuring from 1 to 10 000 base pairs in length, including insertion and deletion events that may be separated by many years, and may not be related to each other in any way. A microindel is defined as an indel that results in a net change of 1 to 50 nucleotides. In coding regions of the genome, unless the length of an indel is a multiple of 3, it will produce a frameshift mutation. For example, a common microindel which results in a frameshift causes Bloom syndrome in the Jewish or Japanese population. Indels can be contrasted with a point mutation. An indel inserts or deletes nucleotides from a sequence, while a point mutation is a form of substitution that ''replaces'' one of the nucleotides without changing the overall number in the DNA. Indels can also be contrasted with Tandem Base Mutations (TBM), which may result from fun ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Single-nucleotide Polymorphism
In genetics, a single-nucleotide polymorphism (SNP ; plural SNPs ) is a germline substitution of a single nucleotide at a specific position in the genome. Although certain definitions require the substitution to be present in a sufficiently large fraction of the population (e.g. 1% or more), many publications do not apply such a frequency threshold. For example, at a specific base position in the human genome, the G nucleotide may appear in most individuals, but in a minority of individuals, the position is occupied by an A. This means that there is a SNP at this specific position, and the two possible nucleotide variations – G or A – are said to be the alleles for this specific position. SNPs pinpoint differences in our susceptibility to a wide range of diseases, for example age-related macular degeneration (a common SNP in the CFH gene is associated with increased risk of the disease) or nonalcoholic fatty liver disease (a SNP in the PNPLA3 gene is associated with inc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transposable Element
A transposable element (TE, transposon, or jumping gene) is a nucleic acid sequence in DNA that can change its position within a genome, sometimes creating or reversing mutations and altering the cell's genetic identity and genome size. Transposition often results in duplication of the same genetic material. Barbara McClintock's discovery of them earned her a Nobel Prize in 1983. Its importance in personalized medicine is becoming increasingly relevant, as well as gaining more attention in data analytics given the difficulty of analysis in very high dimensional spaces. Transposable elements make up a large fraction of the genome and are responsible for much of the mass of DNA in a eukaryotic cell. Although TEs are selfish genetic elements, many are important in genome function and evolution. Transposons are also very useful to researchers as a means to alter DNA inside a living organism. There are at least two classes of TEs: Class I TEs or retrotransposons generally functi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Megabase
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA and RNA. Dictated by specific hydrogen bonding patterns, "Watson–Crick" (or "Watson–Crick–Franklin") base pairs (guanine–cytosine and adenine–thymine) allow the DNA helix to maintain a regular helical structure that is subtly dependent on its nucleotide sequence. The complementary nature of this based-paired structure provides a redundant copy of the genetic information encoded within each strand of DNA. The regular structure and data redundancy provided by the DNA double helix make DNA well suited to the storage of genetic information, while base-pairing between DNA and incoming nucleotides provides the mechanism through which DNA polymerase replicates DNA and RNA polymerase transcribes DNA into RNA. Many DNA-binding proteins ca ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Base Pair
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA and RNA. Dictated by specific hydrogen bonding patterns, "Watson–Crick" (or "Watson–Crick–Franklin") base pairs (guanine–cytosine and adenine–thymine) allow the DNA helix to maintain a regular helical structure that is subtly dependent on its nucleotide sequence. The Complementarity (molecular biology), complementary nature of this based-paired structure provides a redundant copy of the genetic information encoded within each strand of DNA. The regular structure and data redundancy provided by the DNA double helix make DNA well suited to the storage of genetic information, while base-pairing between DNA and incoming nucleotides provides the mechanism through which DNA polymerase replicates DNA and RNA polymerase transcribes DNA in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Kilobase
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA and RNA. Dictated by specific hydrogen bonding patterns, "Watson–Crick" (or "Watson–Crick–Franklin") base pairs (guanine–cytosine and adenine–thymine) allow the DNA helix to maintain a regular helical structure that is subtly dependent on its nucleotide sequence. The complementary nature of this based-paired structure provides a redundant copy of the genetic information encoded within each strand of DNA. The regular structure and data redundancy provided by the DNA double helix make DNA well suited to the storage of genetic information, while base-pairing between DNA and incoming nucleotides provides the mechanism through which DNA polymerase replicates DNA and RNA polymerase transcribes DNA into RNA. Many DNA-binding proteins ca ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |