|
Single Chain Variable Fragment
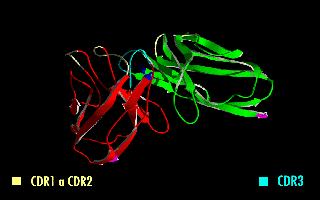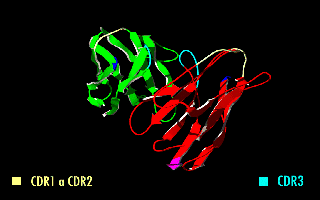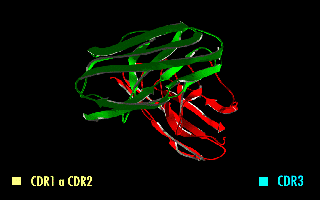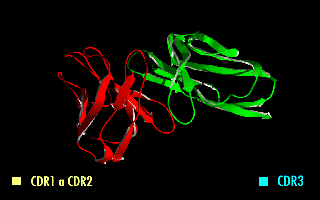
A single-chain variable fragment (scFv) is not actually a fragment of an antibody, but instead is a fusion protein of the variable regions of the heavy (VH) and light chains (VL) of immunoglobulins, connected with a short linker peptide of ten to about 25 amino acids. The linker is usually rich in glycine for flexibility, as well as serine or threonine for solubility, and can either connect the N-terminus of the VH with the C-terminus of the VL, or ''vice versa''. This protein retains the specificity of the original immunoglobulin, despite removal of the constant regions and the introduction of the linker. The image to the right shows how this modification usually leaves the specificity unaltered. These molecules were created to facilitate phage display, where it is highly convenient to express the antigen-binding domain as a single peptide. As an alternative, scFv can be created directly from subcloned heavy and light chains derived from a hybridoma. ScFvs have many uses, e.g. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hybridoma
Hybridoma technology is a method for producing large numbers of identical antibodies (also called monoclonal antibodies). This process starts by injecting a mouse (or other mammal) with an antigen that provokes an immune response. A type of white blood cell, the B cell, produces antibodies that bind to the injected antigen. These antibody producing B-cells are then harvested from the mouse and, in turn, fused with immortal B cell cancer cells, a myeloma, to produce a hybrid cell line called a hybridoma, which has both the antibody-producing ability of the B-cell and the longevity and reproductivity of the myeloma. The hybridomas can be grown in culture, each culture starting with one viable hybridoma cell, producing cultures each of which consists of genetically identical hybridomas which produce one antibody per culture (monoclonal) rather than mixtures of different antibodies (polyclonal). The myeloma cell line that is used in this process is selected for its ability to grow ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bi-specific T-cell Engager
Bi-specific T-cell engagers (BiTEs) are a class of artificial bispecific monoclonal antibodies that are investigated for use as anti-cancer drugs. They direct a host's immune system, more specifically the T cells' cytotoxic activity, against cancer cells. ''BiTE'' is a registered trademark of Micromet AG (fully owned subsidiary of Amgen Inc). BiTEs are fusion proteins consisting of two single-chain variable fragments (scFvs) of different antibodies, or amino acid sequences from four different genes, on a single peptide chain of about 55 kilodaltons. One of the scFvs binds to T cells via the CD3 receptor, and the other to a tumor cell via a tumor specific molecule. Mechanism of action Like other bispecific antibodies, and unlike ordinary monoclonal antibodies, BiTEs form a link between T cells and tumor cells. This causes T cells to exert cytotoxic activity on tumor cells by producing proteins like perforin and granzymes, independently of the presence of MHC I or co-stimulato ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bispecific Antibodies
A bispecific monoclonal antibody (BsMAb, BsAb) is an artificial protein that can simultaneously bind to two different types of antigen or two different epitopes on the same antigen. Naturally occurring antibodies typically only target one antigen. BsAbs can be manufactured in several structural formats. BsAbs can be designed to recruit and activate immune cells, to interfere with receptor signaling and inactivate signaling ligands, and to force association of protein complexes. BsAbs have been explored for cancer immunotherapy, drug delivery, and Alzeimer's disease. Development history The original concept of BsAbs was proposed by Nisonoff and his collaborators in the 1960s, including the first idea of antibody architecture and other findings. In 1975, the problem of producing pure antibodies was solved by the creation of hybridoma technology, and the new era of monoclonal antibodies (MoAbs) came. In 1983, Milstein and Cuello created hybrid-hybridoma (quadroma) technology. In 1988 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Affinity (pharmacology)
In biochemistry and pharmacology, a ligand is a substance that forms a complex with a biomolecule to serve a biological purpose. The etymology stems from ''ligare'', which means 'to bind'. In protein-ligand binding, the ligand is usually a molecule which produces a signal by binding to a site on a target protein. The binding typically results in a change of conformational isomerism (conformation) of the target protein. In DNA-ligand binding studies, the ligand can be a small molecule, ion, or protein which binds to the DNA double helix. The relationship between ligand and binding partner is a function of charge, hydrophobicity, and molecular structure. Binding occurs by intermolecular forces, such as ionic bonds, hydrogen bonds and Van der Waals forces. The association or docking is actually reversible through dissociation. Measurably irreversible covalent bonding between a ligand and target molecule is atypical in biological systems. In contrast to the definition of lig ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dissociation Constant
In chemistry, biochemistry, and pharmacology, a dissociation constant (K_D) is a specific type of equilibrium constant that measures the propensity of a larger object to separate (dissociate) reversibly into smaller components, as when a complex falls apart into its component molecules, or when a salt splits up into its component ions. The dissociation constant is the inverse of the association constant. In the special case of salts, the dissociation constant can also be called an ionization constant. For a general reaction: : A_\mathit B_\mathit \mathit A + \mathit B in which a complex \ce_x \ce_y breaks down into ''x'' A subunits and ''y'' B subunits, the dissociation constant is defined as : K_D = \frac where and ''x'' B''y''are the equilibrium concentrations of A, B, and the complex A''x'' B''y'', respectively. One reason for the popularity of the dissociation constant in biochemistry and pharmacology is that in the frequently en ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polyvalent Single-chain Variable Fragments
Polyvalence or polyvalent may refer to: *Polyvalency (chemistry), chemical species, generally atoms or molecules, which exhibit more than one chemical valence *Polyvalence (music), the musical use of more than one harmonic function of a tonality simultaneously *Polyvalent antibody, a group of antibodies that have affinity for various antigens *Polyvalent logic, a form of many-valued logic or probabilistic logic * Polyvalent vaccine, a vaccine that can vaccinate a person against more than one strain of a disease *Sala Polivalentă (other), various stadiums in Romania commonly translated as Polyvalent Hall *Snake antivenom that contains neutralizing antibodies against two or more species of snakes See also *Bivalence, principle in logic that every declaration is either true or false *Monovalence (other) *Valence (other) Valence or valency may refer to: Science * Valence (chemistry), a measure of an element's combining power with other atoms * Degree (g ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein A
Protein A is a 42 kDa surface protein originally found in the cell wall of the bacteria ''Staphylococcus aureus''. It is encoded by the ''spa'' gene and its regulation is controlled by DNA topology, cellular osmolarity, and a two-component system called ArlS-ArlR. It has found use in biochemical research because of its ability to bind immunoglobulins. It is composed of five homologous Ig-binding domains that fold into a three-helix bundle. Each domain is able to bind proteins from many mammalian species, most notably IgGs. It binds the heavy chain within the Fc region of most immunoglobulins and also within the Fab region in the case of the human VH3 family. Through these interactions in serum, where IgG molecules are bound in the wrong orientation (in relation to normal antibody function), the bacteria disrupts opsonization and phagocytosis. History As a by-product of his work on type-specific staphylococcus antigens, Verwey reported in 1940 that a protein fraction prepar ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein L
Protein L was first isolated from the surface of bacterial species '' Peptostreptococcus magnus'' and was found to bind immunoglobulins through L chain interaction, from which the name was suggested. It consists of 719 amino acid residues. The molecular weight of Protein L purified from the cell walls of '' Peptostreptoccus magnus'' was first estimated as 95kD by SDS-PAGE in the presence of reducing agent 2-mercaptoethanol, while the molecular weight was determined to 76kD by gel chromotography in the presence of 6 M guanidine HCl. Protein L does not contain any interchain disulfide loops, nor does it consist of disulfide-linked subunits. It is an acidic molecule with a pI of 4.0. Unlike Protein A and Protein G, which bind to the Fc region of immunoglobulins ( antibodies), Protein L binds antibodies through light chain interactions. Since no part of the heavy chain is involved in the binding interaction, Protein L binds a wider range of antibody classes than Protein A or G. Prot ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein G
Protein G is an immunoglobulin-binding protein expressed in group C and G Streptococcal bacteria much like Protein A but with differing binding specificities. It is a ~60-kDA (65 kDA for strain G148 and 58 kDa for strain C40) cell surface protein that has found application in purifying antibodies through its binding to the Fab and Fc region. The native molecule also binds albumin, but because serum albumin is a major contaminant of antibody sources, the albumin binding site has been removed from recombinant forms of Protein G. This recombinant Protein G, either labeled with a fluorophore or a single-stranded DNA strand, was used as a replacement for secondary antibodies in immunofluorescence and super-resolution imaging. Other antibody binding proteins In addition to Protein G, other immunoglobulin-binding bacterial proteins such as Protein A, Protein A/G and Protein L are all commonly used to purify, immobilize or detect immunoglobulins. Each of these immunoglobulin-bindin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fc Region
The fragment crystallizable region (Fc region) is the tail region of an antibody that interacts with cell surface receptors called Fc receptors and some proteins of the complement system. This property allows antibodies to activate the immune system. In IgG, IgA and IgD antibody isotypes, the Fc region is composed of two identical protein fragments, derived from the second and third constant domains of the antibody's two heavy chains; IgM and IgE Fc regions contain three heavy chain constant domains (CH domains 2–4) in each polypeptide chain. The Fc regions of IgGs bear a highly conserved N-glycosylation site. Glycosylation of the Fc fragment is essential for Fc receptor-mediated activity. The N-glycans attached to this site are predominantly core- fucosylated diantennary structures of the complex type. In addition, small amounts of these N-glycans also bear bisecting GlcNAc and α-2,6 linked sialic acid residues. The other part of an antibody, called the Fab region, c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Monoclonal Antibodies
A monoclonal antibody (mAb, more rarely called moAb) is an antibody produced from a cell Lineage made by cloning a unique white blood cell. All subsequent antibodies derived this way trace back to a unique parent cell. Monoclonal antibodies can have monovalent affinity, binding only to the same epitope (the part of an antigen that is recognized by the antibody). In contrast, polyclonal antibodies bind to multiple epitopes and are usually made by several different antibody-secreting plasma cell lineages. Bispecific monoclonal antibodies can also be engineered, by increasing the therapeutic targets of one monoclonal antibody to two epitopes. It is possible to produce monoclonal antibodies that specifically bind to virtually any suitable substance; they can then serve to detect or purify it. This capability has become an investigative tool in biochemistry, molecular biology, and medicine. Monoclonal antibodies are being used on a clinical level for both the diagnosis and therap ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |