|
Svedberg
A Svedberg unit or svedberg (symbol S, sometimes Sv) is a non- SI metric unit for sedimentation coefficients. The Svedberg unit offers a measure of a particle's size indirectly based on its sedimentation rate under acceleration (i.e. how fast a particle of given size and shape settles to the bottom of a solution). The svedberg is a measure of time, defined as exactly 10−13 seconds (100 fs). For biological macromolecules and cell organelles like ribosomes, the sedimentation rate is typically measured as the rate of travel in a centrifuge tube subjected to high g-force. The svedberg (S) is distinct from the SI unit sievert or the non-SI unit sverdrup, which also use the symbol Sv. Naming The unit is named after the Swedish chemist Theodor Svedberg (1884–1971), winner of the 1926 Nobel Prize in chemistry for his work on disperse systems, colloids and his invention of the ultracentrifuge. Factors The Svedberg coefficient is a nonlinear function. A particle's mas ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Theodor Svedberg
Theodor Svedberg (30 August 1884 – 25 February 1971) was a Swedish chemist and Nobel laureate for his research on colloids and proteins using the ultracentrifuge. Svedberg was active at Uppsala University from the mid 1900s to late 1940s. While at Uppsala, Svedberg started as a docent before becoming the university's physical chemistry head in 1912. After leaving Uppsala in 1949, Svedberg was in charge of the Gustaf Werner Institute until 1967. Apart from his 1926 Nobel Prize, Svedberg was named a Foreign Member of the Royal Society in 1944 and became part of the National Academy of Sciences in 1945. Early life and education Svedberg was born in Valbo, Sweden on 30 August 1884. He was the son of Augusta Alstermark and Elias Svedberg. Growing up, Svedberg enjoyed botany and other branches of science. While in grammar school, Svedberg conducted individual laboratorial research and performed scientific demonstrations. For his post-secondary education, Svedberg entered a c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ultracentrifuge
An ultracentrifuge is a centrifuge optimized for spinning a rotor at very high speeds, capable of generating acceleration as high as (approx. ). There are two kinds of ultracentrifuges, the preparative and the analytical ultracentrifuge. Both classes of instruments find important uses in molecular biology, biochemistry, and polymer science.Susan R. Mikkelsen & Eduardo Cortón. Bioanalytical Chemistry, Ch. 13. Centrifugation Methods. John Wiley & Sons, Mar 4, 2004, pp. 247-267. History In 1924 Theodor Svedberg built a centrifuge capable of generating 7,000 g (at 12,000 rpm), and called it the ultracentrifuge, to juxtapose it with the Ultramicroscope that had been developed previously. In 1925-1926 Svedberg constructed a new ultracentrifuge that permitted fields up to 100,000 g (42,000 rpm). Modern ultracentrifuges are typically classified as allowing greater than 100,000 g. Svedberg won the Nobel Prize in Chemistry in 1926 for his research on colloids and proteins using the ultrac ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribosome
Ribosomes ( ) are macromolecular machines, found within all cells, that perform biological protein synthesis (mRNA translation). Ribosomes link amino acids together in the order specified by the codons of messenger RNA (mRNA) molecules to form polypeptide chains. Ribosomes consist of two major components: the small and large ribosomal subunits. Each subunit consists of one or more ribosomal RNA (rRNA) molecules and many ribosomal proteins (RPs or r-proteins). The ribosomes and associated molecules are also known as the ''translational apparatus''. Overview The sequence of DNA that encodes the sequence of the amino acids in a protein is transcribed into a messenger RNA chain. Ribosomes bind to messenger RNAs and use their sequences for determining the correct sequence of amino acids to generate a given protein. Amino acids are selected and carried to the ribosome by transfer RNA, transfer RNA (tRNA) molecules, which enter the ribosome and bind to the messenger RNA chain vi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
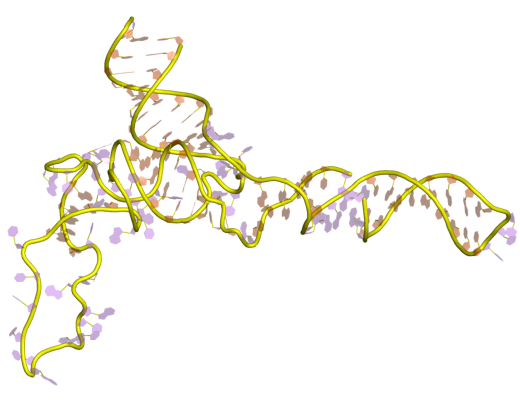
5S Ribosomal RNA
The 5S ribosomal RNA (5S rRNA) is an approximately 120 nucleotide-long ribosomal RNA molecule with a mass of 40 kDa. It is a structural and functional component of the large subunit of the ribosome in all domains of life (bacteria, archaea, and eukaryotes), with the exception of mitochondrial ribosomes of fungi and animals. The designation 5S refers to the molecule's sedimentation velocity in an ultracentrifuge, which is measured in Svedberg units (S). Biosynthesis In prokaryotes, the 5S rRNA gene is typically located in the rRNA operons downstream of the small and the large subunit rRNA, and co-transcribed into a polycistronic precursor. A particularity of eukaryotic nuclear genomes is the occurrence of multiple 5S rRNA gene copies (5S rDNA) clustered in tandem repeats, with copy number varying from species to species. Eukaryotic 5S rRNA is synthesized by RNA polymerase III, whereas other eukaryotic rRNAs are cleaved from a 45S precursor transcribed by RNA polymerase I. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
16S Ribosomal RNA
16 S ribosomal RNA (or 16 S rRNA) is the RNA component of the 30S subunit of a prokaryotic ribosome ( SSU rRNA). It binds to the Shine-Dalgarno sequence and provides most of the SSU structure. The genes coding for it are referred to as 16S rRNA gene and are used in reconstructing phylogenies, due to the slow rates of evolution of this region of the gene. Carl Woese and George E. Fox were two of the people who pioneered the use of 16S rRNA in phylogenetics in 1977. Multiple sequences of the 16S rRNA gene can exist within a single bacterium. Functions * Like the large (23S) ribosomal RNA, it has a structural role, acting as a scaffold defining the positions of the ribosomal proteins. * The 3-end contains the anti- Shine-Dalgarno sequence, which binds upstream to the AUG start codon on the mRNA. The 3-end of 16S RNA binds to the proteins S1 and S21 which are known to be involved in initiation of protein synthesis * Interacts with 23S, aiding in the binding of the two ribo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RRNA
Ribosomal ribonucleic acid (rRNA) is a type of non-coding RNA which is the primary component of ribosomes, essential to all cells. rRNA is a ribozyme which carries out protein synthesis in ribosomes. Ribosomal RNA is transcribed from ribosomal DNA (rDNA) and then bound to ribosomal proteins to form small and large ribosome subunits. rRNA is the physical and mechanical factor of the ribosome that forces transfer RNA (tRNA) and messenger RNA (mRNA) to process and translate the latter into proteins. Ribosomal RNA is the predominant form of RNA found in most cells; it makes up about 80% of cellular RNA despite never being translated into proteins itself. Ribosomes are composed of approximately 60% rRNA and 40% ribosomal proteins by mass. Structure Although the primary structure of rRNA sequences can vary across organisms, base-pairing within these sequences commonly forms stem-loop configurations. The length and position of these rRNA stem-loops allow them to create thre ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sedimentation Coefficient
The sedimentation coefficient () of a particle characterizes its sedimentation during centrifugation. It is defined as the ratio of a particle's sedimentation velocity to the applied acceleration causing the sedimentation. : s = \frac The sedimentation speed is also the terminal velocity. It is constant because the force applied to a particle by gravity or by a centrifuge (typically in multiples of tens of thousands of gravities in an ultracentrifuge) is balanced by the viscous resistance (or "drag") of the fluid (normally water) through which the particle is moving. The applied acceleration can be either the gravitational acceleration , or more commonly the centrifugal acceleration . In the latter case, is the angular velocity of the rotor and is the distance of a particle to the rotor axis ( radius). The viscous resistance for a spherical particle is given by Stokes' law: , where is the viscosity of the medium, is the radius of the particle and is the velocity ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Solution (chemistry)
In chemistry, a solution is a special type of homogeneous mixture composed of two or more substances. In such a mixture, a solute is a substance dissolved in another substance, known as a solvent. If the attractive forces between the solvent and solute particles are greater than the attractive forces holding the solute particles together, the solvent particles pull the solute particles apart and surround them. These surrounded solute particles then move away from the solid solute and out into the solution. The mixing process of a solution happens at a scale where the effects of chemical polarity are involved, resulting in interactions that are specific to solvation. The solution usually has the state of the solvent when the solvent is the larger fraction of the mixture, as is commonly the case. One important parameter of a solution is the concentration, which is a measure of the amount of solute in a given amount of solution or solvent. The term " aqueous solution" is used ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Settling
Settling is the process by which particulates move towards the bottom of a liquid and form a sediment. Particles that experience a force, either due to gravity or due to centrifugal motion will tend to move in a uniform manner in the direction exerted by that force. For gravity settling, this means that the particles will tend to fall to the bottom of the vessel, forming sludge or slurry at the vessel base. Settling is an important operation in many applications, such as mining, wastewater and drinking water treatment, biological science, space propellant reignition, and scooping. Physics For settling particles that are considered individually, i.e. dilute particle solutions, there are two main forces enacting upon any particle. The primary force is an applied force, such as gravity, and a drag force that is due to the motion of the particle through the fluid. The applied force is usually not affected by the particle's velocity, whereas the drag force is a function of th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribosomal Protein
A ribosomal protein (r-protein or rProtein) is any of the proteins that, in conjunction with rRNA, make up the ribosomal subunits involved in the cellular process of translation. ''E. coli'', other bacteria and Archaea have a 30S small subunit and a 50S large subunit, whereas humans and yeasts have a 40S small subunit and a 60S large subunit. Equivalent subunits are frequently numbered differently between bacteria, Archaea, yeasts and humans. A large part of the knowledge about these organic molecules has come from the study of '' E. coli'' ribosomes. All ribosomal proteins have been isolated and many specific antibodies have been produced. These, together with electronic microscopy and the use of certain reactives, have allowed for the determination of the topography of the proteins in the ribosome. More recently, a near-complete (near)atomic picture of the ribosomal proteins is emerging from the latest high-resolution cryo-EM data (including ). Conservation Ribosomal p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Metric Units
Metric units are units based on the metre, gram or second and decimal (power of ten) multiples or sub-multiples of these. The most widely used examples are the units of the International System of Units (SI). By extension they include units of electromagnetism from the CGS and SI units systems, and other units for which use of SI prefixes has become the norm. Other unit systems using metric units include: * International System of Electrical and Magnetic Units * Metre–tonne–second (MTS) system of units * MKS system of units (metre, kilogram, second) Metric units that are part of the SI The first group of metric units are those that are at present defined as units within the International System of Units (SI). In its most restrictive interpretation, this is what may be meant when the term ''metric unit'' is used. The SI defines 30 named units and associated symbols: * The unit one (1) is the unit of a quantity of dimension one. * The second (s) is the unit of time. * ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |