|
Response Regulator
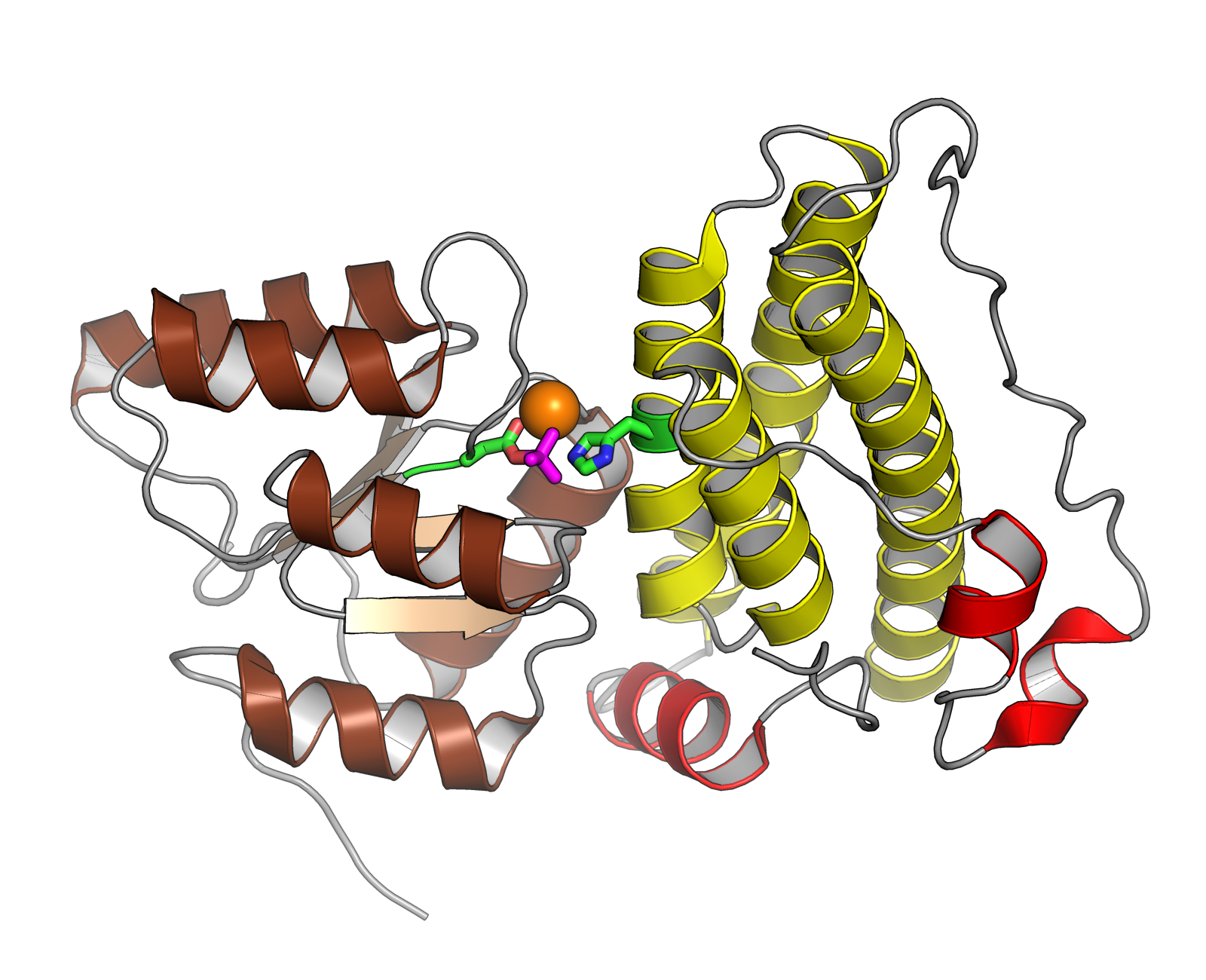
A response regulator is a protein that mediates a cell's response to changes in its environment as part of a two-component regulatory system. Response regulators are coupled to specific histidine kinases which serve as sensors of environmental changes. Response regulators and histidine kinases are two of the most common gene families in bacteria, where two-component signaling systems are very common; they also appear much more rarely in the genomes of some archaea, yeasts, filamentous fungi, and plants. Two-component systems are not found in metazoans. Function Response regulator proteins typically consist of a receiver domain and one or more effector domains, although in some cases they possess only a receiver domain and exert their effects through protein-protein interactions. In two-component signaling, a histidine kinase responds to environmental changes by autophosphorylation on a histidine residue, following which the response regulator receiver domain catalyzes transfer ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CheY , Canadian radio station
{{dab ...
Chey may refer to: People Given name * Chey Chettha I (1575–1595), Cambodian king * Chey Chettha II, Cambodian king * Chey Chettha III (1639–1673), Cambodian king * Chey Chettha IV (1656–1725), Cambodian king * Chey Chettha V (1709–1755), Cambodian king * Chey Dunkley (born 1992), English football player Surname * Chey Tae-won Tae-won (born 1960), South Korea billionaire businessman * Timothy A. Chey, American film producer, writer and director Places * Chey, Deux-Sèvres, Nouvelle-Aquitaine, France * Chey, Kampong Svay, Cambodia * Chey Chouk Commune, Cambodia * Chey Saen District, Cambodia Other * CHEY-FM CHEY-FM is a French-language radio station located in Trois-Rivières, Quebec, Canada. Owned and operated by Bell Media, it broadcasts on 94.7 MHz using a directional antenna with an average effective radiated power of 20,600 watts and a peak ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Autophosphorylation
Autophosphorylation is a type of post-translational modification of proteins. It is generally defined as the phosphorylation of the kinase by itself. In eukaryotes, this process occurs by the addition of a phosphate group to serine, threonine or tyrosine residues within protein kinases, normally to regulate the catalytic activity.Petsko, GA and Ringe, D 2009, 'Protein Structure and Function', Oxford University Press Inc., New York, U.S.A Autophosphorylation may occur when a kinases' own active site catalyzes the phosphorylation reaction (cis autophosphorylation), or when another kinase of the same type provides the active site that carries out the chemistry (trans autophosphorylation). The latter often occurs when kinase molecules dimerize. In general, the phosphate groups introduced are gamma phosphates from nucleoside triphosphates, most commonly ATP. Function Protein kinases, many of which are regulated by autophosphorylation, are vital in controlling the cellular prolifer ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
EnvZ/OmpR Two-component System
EnvZ/OmpR is a two-component regulatory system widely distributed in bacteria and particularly well characterized in ''Escherichia coli''. Its function is in osmoregulation, responding to changes in environmental osmolality by regulating the expression of the outer membrane porins OmpF and OmpC. EnvZ is a histidine kinase which also possesses a cytoplasmic osmosensory domain, and OmpR is its corresponding response regulator protein. Overview EnvZ is a sensor-transmitter that spans the inner cytoplasmic membrane and has historically been divided into two domains, the sensory and the transmitter domain. The protein is composed of a short N-terminal tail in the periplasm, two transmembrane regions with an intervening periplasmic loop, and a cytoplasmic domain containing the autophosphorylated histidine residue, which is located in a dimeric four-helix bundle. Variations in osmolality trigger conformational changes in EnvZ which have been localized to the cytoplasmic four-helix bu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Helix-turn-helix
Helix-turn-helix is a DNA-binding protein (DBP). The helix-turn-helix (HTH) is a major structural motif capable of binding DNA. Each monomer incorporates two α helices, joined by a short strand of amino acids, that bind to the major groove of DNA. The HTH motif occurs in many proteins that regulate gene expression. It should not be confused with the helix–loop–helix motif. Discovery The discovery of the helix-turn-helix motif was based on similarities between several genes encoding transcription regulatory proteins from bacteriophage lambda and ''Escherichia coli'': Cro, CAP, and λ repressor, which were found to share a common 20–25 amino acid sequence that facilitates DNA recognition. Function The helix-turn-helix motif is a DNA-binding motif. The recognition and binding to DNA by helix-turn-helix proteins is done by the two α helices, one occupying the N-terminal end of the motif, the other at the C-terminus. In most cases, such as in the Cro repressor, the second ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription Factor
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The function of TFs is to regulate—turn on and off—genes in order to make sure that they are expressed in the desired cells at the right time and in the right amount throughout the life of the cell and the organism. Groups of TFs function in a coordinated fashion to direct cell division, cell growth, and cell death throughout life; cell migration and organization (body plan) during embryonic development; and intermittently in response to signals from outside the cell, such as a hormone. There are up to 1600 TFs in the human genome. Transcription factors are members of the proteome as well as regulome. TFs work alone or with other proteins in a complex, by promoting (as an activator), or blocking (as a repressor) the recruitment of RNA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription (biology)
Transcription is the process of copying a segment of DNA into RNA. The segments of DNA transcribed into RNA molecules that can encode proteins are said to produce messenger RNA (mRNA). Other segments of DNA are copied into RNA molecules called non-coding RNAs (ncRNAs). mRNA comprises only 1–3% of total RNA samples. Less than 2% of the human genome can be transcribed into mRNA ( Human genome#Coding vs. noncoding DNA), while at least 80% of mammalian genomic DNA can be actively transcribed (in one or more types of cells), with the majority of this 80% considered to be ncRNA. Both DNA and RNA are nucleic acids, which use base pairs of nucleotides as a complementary language. During transcription, a DNA sequence is read by an RNA polymerase, which produces a complementary, antiparallel RNA strand called a primary transcript. Transcription proceeds in the following general steps: # RNA polymerase, together with one or more general transcription factors, binds to promoter ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA-binding Protein
DNA-binding proteins are proteins that have DNA-binding domains and thus have a specific or general affinity for DNA#Base pairing, single- or double-stranded DNA. Sequence-specific DNA-binding proteins generally interact with the major groove of B-DNA, because it exposes more functional groups that identify a base pair. However, there are some known minor groove DNA-binding ligands such as netropsin, distamycin, Hoechst 33258, pentamidine, DAPI and others. Examples DNA-binding proteins include transcription factors which Gene modulation, modulate the process of transcription, various polymerases, nucleases which cleave DNA molecules, and histones which are involved in chromosome packaging and transcription in the cell nucleus. DNA-binding proteins can incorporate such domains as the zinc finger, the helix-turn-helix, and the leucine zipper (among many others) that facilitate binding to nucleic acid. There are also more unusual examples such as TAL effector, transcription activa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phosphatase
In biochemistry, a phosphatase is an enzyme that uses water to cleave a phosphoric acid Ester, monoester into a phosphate ion and an Alcohol (chemistry), alcohol. Because a phosphatase enzyme catalysis, catalyzes the hydrolysis of its Substrate (chemistry), substrate, it is a subcategory of hydrolases. Phosphatase enzymes are essential to many biological functions, because phosphorylation (e.g. by protein kinases) and dephosphorylation (by phosphatases) serve diverse roles in cell growth, cellular regulation and cell signaling, signaling. Whereas phosphatases remove phosphate groups from molecules, kinases catalyze the transfer of phosphate groups to molecules from Adenosine triphosphate, ATP. Together, kinases and phosphatases direct a form of post-translational modification that is essential to the cell's regulatory network. Phosphatase enzymes are not to be confused with phosphorylase enzymes, which catalyze the transfer of a phosphate group from hydrogen phosphate to an acce ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histidine Phosphotransfer Protein
Histidine phosphotransfer domains and histidine phosphotransferases (both often abbreviated HPt) are protein domains involved in the "phosphorelay" form of two-component regulatory systems. These proteins possess a phosphorylatable histidine residue and are responsible for transferring a phosphoryl group from an aspartate residue on an intermediate "receiver" domain, typically part of a hybrid histidine kinase, to an aspartate on a final response regulator. Function In orthodox two-component signaling, a histidine kinase protein autophosphorylates on a histidine residue in response to an extracellular signal, and the phosphoryl group is subsequently transferred to an aspartate residue on the receiver domain of a response regulator. In phosphorelays, the "hybrid" histidine kinase contains an internal aspartate-containing receiver domain to which the phosphoryl group is transferred, after which an HPt protein containing a phosphorylatable histidine receives the phosphoryl group ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Oligomer
In chemistry and biochemistry, an oligomer () is a molecule that consists of a few repeating units which could be derived, actually or conceptually, from smaller molecules, monomers.Quote: ''Oligomer molecule: A molecule of intermediate relative molecular mass, the structure of which essentially comprises a small plurality of units derived, actually or conceptually, from molecules of lower relative molecular mass.'' The name is composed of Greek elements '' oligo-'', "a few" and '' -mer'', "parts". An adjective form is ''oligomeric''. The oligomer concept is contrasted to that of a polymer, which is usually understood to have a large number of units, possibly thousands or millions. However, there is no sharp distinction between these two concepts. One proposed criterion is whether the molecule's properties vary significantly with the removal of one or a few of the units. An oligomer with a specific number of units is referred to by the Greek prefix denoting that number, wi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Allosteric
In biochemistry, allosteric regulation (or allosteric control) is the regulation of an enzyme by binding an effector molecule at a site other than the enzyme's active site. The site to which the effector binds is termed the ''allosteric site'' or ''regulatory site''. Allosteric sites allow effectors to bind to the protein, often resulting in a conformational change and/or a change in protein dynamics. Effectors that enhance the protein's activity are referred to as ''allosteric activators'', whereas those that decrease the protein's activity are called ''allosteric inhibitors''. Allosteric regulations are a natural example of control loops, such as feedback from downstream products or feedforward from upstream substrates. Long-range allostery is especially important in cell signaling. Allosteric regulation is also particularly important in the cell's ability to adjust enzyme activity. The term ''allostery'' comes from the Ancient Greek ''allos'' (), "other", and ''stereos' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription (genetics)
Transcription is the process of copying a segment of DNA into RNA. The segments of DNA transcribed into RNA molecules that can encode proteins are said to produce messenger RNA (mRNA). Other segments of DNA are copied into RNA molecules called non-coding RNAs (ncRNAs). mRNA comprises only 1–3% of total RNA samples. Less than 2% of the human genome can be transcribed into mRNA ( Human genome#Coding vs. noncoding DNA), while at least 80% of mammalian genomic DNA can be actively transcribed (in one or more types of cells), with the majority of this 80% considered to be ncRNA. Both DNA and RNA are nucleic acids, which use base pairs of nucleotides as a complementary language. During transcription, a DNA sequence is read by an RNA polymerase, which produces a complementary, antiparallel RNA strand called a primary transcript. Transcription proceeds in the following general steps: # RNA polymerase, together with one or more general transcription factors, binds to promoter DNA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |