|
Repressilator
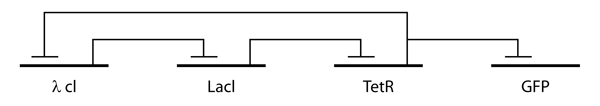
The repressilator is a genetic regulatory network consisting of at least one feedback loop with at least three genes, each expressing a protein that represses the next gene in the loop. In biological research, repressilators have been used to build cellular models and understand cell function. There are both artificial and naturally-occurring repressilators. Recently, the naturally-occurring repressilator clock gene circuit in '' Arabidopsis thaliana'' (''A. thaliana'') and mammalian systems have been studied. Artificial Repressilators Artificial repressilators were first engineered by Michael Elowitz and Stanislas Leibler in 2000, complementing other research projects studying simple systems of cell components and function. In order to understand and model the design and cellular mechanisms that confers a cell’s function, Elowitz and Leibler created an artificial network consisting of a loop with three transcriptional repressors. This network was designed from scratch to exhi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Repressilator GRN
The repressilator is a genetic regulatory network consisting of at least one feedback loop with at least three genes, each expressing a protein that represses the next gene in the loop. In biological research, repressilators have been used to build cellular models and understand cell function. There are both artificial and naturally-occurring repressilators. Recently, the naturally-occurring repressilator clock gene circuit in ''Arabidopsis thaliana'' (''A. thaliana'') and mammalian systems have been studied. Artificial Repressilators Artificial repressilators were first engineered by Michael Elowitz and Stanislas Leibler in 2000, complementing other research projects studying simple systems of cell components and function. In order to understand and model the design and cellular mechanisms that confers a cell’s function, Elowitz and Leibler created an artificial network consisting of a loop with three transcriptional repressors. This network was designed from scratch to exhibi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Repressilator Plasmid
The repressilator is a genetic regulatory network consisting of at least one feedback loop with at least three genes, each expressing a protein that represses the next gene in the loop. In biological research, repressilators have been used to build cellular models and understand cell function. There are both artificial and naturally-occurring repressilators. Recently, the naturally-occurring repressilator clock gene circuit in ''Arabidopsis thaliana'' (''A. thaliana'') and mammalian systems have been studied. Artificial Repressilators Artificial repressilators were first engineered by Michael Elowitz and Stanislas Leibler in 2000, complementing other research projects studying simple systems of cell components and function. In order to understand and model the design and cellular mechanisms that confers a cell’s function, Elowitz and Leibler created an artificial network consisting of a loop with three transcriptional repressors. This network was designed from scratch to exhibi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TOC1 (gene)
Timing of CAB expression 1 is a protein that in '' Arabidopsis thaliana'' is encoded by the TOC1 gene. TOC1 is also known as two-component response regulator-like APRR1. TOC1 was the first plant gene that, when mutated, yielded a circadian phenotype. It codes for the transcription factor TOC1, which affects the period of plants' circadian rhythms: built-in, malleable oscillations that repeat every 24 hours. The gene codes for a transcriptional repressor, TOC1, one of five pseudo-response regulators (PRR) that mediate the period of the circadian clock in plants. The TOC1 protein is involved in the clock's evening loop, which is a repressilator that directly inhibits transcription of morning loop genes LHY and CCA1. Toc1 gene is expressed in most plant structures and cells, and has its locus on chromosome 5. Historical context Discovery The TOC1 gene was initially discovered by Prof. Andrew Millar and colleagues in 1995 while Millar was a graduate student. Millar develo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Circadian Clock Associated 1
Circadian Clock Associated 1 (CCA1) is a gene that is central to the Circadian clock, circadian oscillator of angiosperms. It was first identified in ''Arabidopsis thaliana'' in 1993. CCA1 interacts with LHY and TOC1 (gene), TOC1 to form the core of the oscillator system. CCA1 expression peaks at dawn. Loss of CCA1 function leads to a shortened period in the expression of many other genes. Discovery CCA1 was first identified in ''Arabidopsis thaliana'' by Elaine M. Tobin’s lab in University of California, Los Angeles, UCLA in 1993. Tobin’s lab was studying promoter fragments that contribute to light regulation of light-harvesting Chlorophyll A/B Binding Protein (LHCB), and noticed DNA-binding activity that had affinity for a specific light-responsive fragment of the LHCB promoter. This DNA-binding activity was designated as CA-1 because the binding is mostly to cytosine and adenine-rich sequences. They found that this binding activity is necessary for phytochrome response. T ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genetic Regulatory Network
A gene (or genetic) regulatory network (GRN) is a collection of molecular regulators that interact with each other and with other substances in the cell to govern the gene expression levels of mRNA and proteins which, in turn, determine the function of the cell. GRN also play a central role in morphogenesis, the creation of body structures, which in turn is central to evolutionary developmental biology (evo-devo). The regulator can be DNA, RNA, protein or any combination of two or more of these three that form a complex, such as a specific sequence of DNA and a transcription factor to activate that sequence. The interaction can be direct or indirect (through transcribed RNA or translated protein). In general, each mRNA molecule goes on to make a specific protein (or set of proteins). In some cases this protein will be Protein#Structural proteins, structural, and will accumulate at the cell membrane or within the cell to give it particular structural properties. In other cases th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Lambda Phage
''Enterobacteria phage λ'' (lambda phage, coliphage λ, officially ''Escherichia virus Lambda'') is a bacterial virus, or bacteriophage, that infects the bacterial species ''Escherichia coli'' (''E. coli''). It was discovered by Esther Lederberg in 1950. The wild type of this virus has a temperate life cycle that allows it to either reside within the genome of its host through lysogeny or enter into a lytic phase, during which it kills and lyses the cell to produce offspring. Lambda strains, mutated at specific sites, are unable to lysogenize cells; instead, they grow and enter the lytic cycle after superinfecting an already lysogenized cell. The phage particle consists of a head (also known as a capsid), a tail, and tail fibers (see image of virus below). The head contains the phage's double-strand linear DNA genome. During infection, the phage particle recognizes and binds to its host, ''E. coli'', causing DNA in the head of the phage to be ejected through the tail into the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Period (gene)
Period (per) is a gene located on the X chromosome of ''Drosophila melanogaster''. Oscillations in levels of both ''per'' transcript and its corresponding protein PER have a period of approximately 24 hours and together play a central role in the molecular mechanism of the ''Drosophila'' biological clock driving circadian rhythms in eclosion and locomotor activity. Mutations in the per gene can shorten (''perS''), lengthen (''perL''), and even abolish (''per0'') the period of the circadian rhythm. Discovery The period gene and three mutants (''perS'', ''perL'', and ''per0'') were isolated in an EMS mutagenesis screen by Ronald Konopka and Seymour Benzer in 1971. The ''perS'', ''perL'', and ''per0'' mutations were found to not complement each other, so it was concluded that the three phenotypes were due to mutations in the same gene. The discovery of mutants that altered the period of circadian rhythms in eclosion and locomotor activity (''perS'' and ''perL'') indicated the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Photoperiodism
Photoperiodism is the physiological reaction of organisms to the length of night or a dark period. It occurs in plants and animals. Plant photoperiodism can also be defined as the developmental responses of plants to the relative lengths of light and dark periods. They are classified under three groups according to the photoperiods: short-day plants, long-day plants, and day-neutral plants. Plants Many flowering plants (angiosperms) use a photoreceptor protein, such as phytochrome or cryptochrome, to sense seasonal changes in night length, or photoperiod, which they take as signals to flower. In a further subdivision, ''obligate'' photoperiodic plants absolutely require a long or short enough night before flowering, whereas ''facultative'' photoperiodic plants are more likely to flower under one condition. Phytochrome comes in two forms: Pr and Pfr. Red light (which is present during the day) converts phytochrome to its active form (pfr). This then triggers the plant to grow. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hypocotyl
The hypocotyl (short for "hypocotyledonous stem", meaning "below seed leaf") is the stem of a germinating seedling, found below the cotyledons (seed leaves) and above the radicle (root). Eudicots As the plant embryo grows at germination, it sends out a shoot called a radicle that becomes the primary root, and then penetrates down into the soil. After emergence of the radicle, the hypocotyl emerges and lifts the growing tip (usually including the seed coat) above the ground, bearing the embryonic leaves (called cotyledons), and the plumule that gives rise to the first true leaves. The hypocotyl is the primary organ of extension of the young plant and develops into the stem. Monocots The early development of a monocot seedling like cereals and other grasses is somewhat different. A structure called the coleoptile, essentially a part of the ''cotyledon'', protects the young stem and plumule as growth pushes them up through the soil. A mesocotyl—that part of the young plant that li ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Late Elongated Hypocotyl
The Late Elongated Hypocotyl gene (LHY), is an oscillating gene found in plants that functions as part of their circadian clock. LHY encodes components of mutually regulatory negative feedback loops with Circadian Clock Associated 1 (CCA1) in which overexpression of either results in dampening of both of their expression. This negative feedback loop affects the rhythmicity of multiple outputs creating a daytime protein complex. LHY was one of the first genes identified in the plant clock, along with TOC1 and CCA1. LHY and CCA1 have similar patterns of expression, which is capable of being induced by light.Lu, S. X. , and S. M., Andronis. "CIRCADIAN CLOCK ASSOCIATED1 and LATE ELONGATED HYPOCOTYL Function Synergistically in the Circadian Clock of Arabidopsis" Plant Physiology Vol. 150. (2009): 834–843. Single loss-of-function mutants in both genes result in seemingly identical phenotypes, but LHY cannot fully rescue the rhythm when CCA1 is absent, indicating that they may only be p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phytochrome
Phytochromes are a class of photoreceptor in plants, bacteria and fungi used to detect light. They are sensitive to light in the red and far-red region of the visible spectrum and can be classed as either Type I, which are activated by far-red light, or Type II that are activated by red light. Recent advances have suggested that phytochromes also act as temperature sensors, as warmer temperatures enhance their de-activation. All of these factors contribute to the plant's ability to germinate. Phytochromes control many aspects of plant development. They regulate the germination of seeds (photoblasty), the synthesis of chlorophyll, the elongation of seedlings, the size, shape and number and movement of leaves and the timing of flowering in adult plants. Phytochromes are widely expressed across many tissues and developmental stages. Other plant photoreceptors include cryptochromes and phototropins, which respond to blue and ultraviolet-A light and UVR8, which is sensitive to u ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cryptochrome
Cryptochromes (from the Greek κρυπτός χρώμα, "hidden colour") are a class of flavoproteins found in plants and animals that are sensitive to blue light. They are involved in the circadian rhythms and the sensing of magnetic fields in a number of species. The name ''cryptochrome'' was proposed as a ''portmanteau'' combining the '' chromatic'' nature of the photoreceptor, and the ''cryptogamic'' organisms on which many blue-light studies were carried out. The two genes ''Cry1'' and ''Cry2'' code the two cryptochrome proteins CRY1 and CRY2. In insects and plants, CRY1 regulates the circadian clock in a light-dependent fashion, whereas in mammals, CRY1 and CRY2 act as light-independent inhibitors of CLOCK-BMAL1 components of the circadian clock. In plants, blue-light photoreception can be used to cue developmental signals. Besides chlorophylls, cryptochromes are the only proteins known to form photoinduced radical-pairs ''in vivo''. These appear to enable some animal ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |