|
Rolling Circle
Rolling circle replication (RCR) is a process of unidirectional nucleic acid replication that can rapidly synthesize multiple copies of circular molecules of DNA or RNA, such as plasmids, the genomes of bacteriophages, and the circular RNA genome of viroids. Some eukaryotic viruses also replicate their DNA or RNA via the rolling circle mechanism. As a simplified version of natural rolling circle replication, an isothermal DNA amplification technique, rolling circle amplification was developed. The RCA mechanism is widely used in molecular biology and biomedical nanotechnology, especially in the field of biosensing (as a method of signal amplification). Circular DNA replication Rolling circle DNA replication is initiated by an initiator protein encoded by the plasmid or bacteriophage DNA, which nicks one strand of the double-stranded, circular DNA molecule at a site called the double-strand origin, or DSO. The initiator protein remains bound to the 5' phosphate end of the ni ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Rolling Circle
Rolling circle replication (RCR) is a process of unidirectional nucleic acid replication that can rapidly synthesize multiple copies of circular molecules of DNA or RNA, such as plasmids, the genomes of bacteriophages, and the circular RNA genome of viroids. Some eukaryotic viruses also replicate their DNA or RNA via the rolling circle mechanism. As a simplified version of natural rolling circle replication, an isothermal DNA amplification technique, rolling circle amplification was developed. The RCA mechanism is widely used in molecular biology and biomedical nanotechnology, especially in the field of biosensing (as a method of signal amplification). Circular DNA replication Rolling circle DNA replication is initiated by an initiator protein encoded by the plasmid or bacteriophage DNA, which nicks one strand of the double-stranded, circular DNA molecule at a site called the double-strand origin, or DSO. The initiator protein remains bound to the 5' phosphate end of the ni ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Polymerase I
DNA polymerase I (or Pol I) is an enzyme that participates in the process of prokaryotic DNA replication. Discovered by Arthur Kornberg in 1956, it was the first known DNA polymerase (and the first known of any kind of polymerase). It was initially characterized in '' E. coli'' and is ubiquitous in prokaryotes. In ''E. coli'' and many other bacteria, the gene that encodes Pol I is known as ''polA''. The ''E. coli'' Pol I enzyme is composed of 928 amino acids, and is an example of a processive enzyme — it can sequentially catalyze multiple polymerisation steps without releasing the single-stranded template. The physiological function of Pol I is mainly to support repair of damaged DNA, but it also contributes to connecting Okazaki fragments by deleting RNA primers and replacing the ribonucleotides with DNA. Discovery In 1956, Arthur Kornberg and colleagues discovered Pol I by using ''Escherichia coli'' (''E. coli'') extracts to develop a DNA synthesis assay. The scientist ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Rolling Circle Replication Of RNA Viroids
Rolling is a Motion (physics)#Types of motion, type of motion that combines rotation (commonly, of an Axial symmetry, axially symmetric object) and Translation (geometry), translation of that object with respect to a surface (either one or the other moves), such that, if ideal conditions exist, the two are in contact with each other without sliding (motion), sliding. Rolling where there is no sliding is referred to as ''pure rolling''. By definition, there is no sliding when there is a frame of reference in which all points of contact on the rolling object have the same velocity as their counterparts on the surface on which the object rolls; in particular, for a frame of reference in which the rolling plane is at rest (see animation), the instantaneous velocity of all the points of contact (e.g., a generating line segment of a cylinder) of the rolling object is zero. In practice, due to small deformations near the contact area, some sliding and energy dissipation occurs. Neverthe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enterobacteria Phage T4
Escherichia virus T4 is a species of bacteriophages that infect ''Escherichia coli'' bacteria. It is a double-stranded DNA virus in the subfamily '' Tevenvirinae'' from the family Myoviridae. T4 is capable of undergoing only a lytic lifecycle and not the lysogenic lifecycle. The species was formerly named T-even bacteriophage, a name which also encompasses, among other strains (or isolates), Enterobacteria phage T2, Enterobacteria phage T4 and Enterobacteria phage T6. Use in research Dating back to the 1940s and continuing today, T-even phages are considered the best studied model organisms. Model organisms are usually required to be simple with as few as five genes. Yet, T-even phages are in fact among the largest and highest complexity virus, in which these phage's genetic information is made up of around 300 genes. Coincident with their complexity, T-even viruses were found to have the unusual base hydroxymethylcytosine (HMC) in place of the nucleic acid base cytosine. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Geminiviridae
''Geminiviridae'' is a family of plant viruses that encode their genetic information on a circular genome of single-stranded (ss) DNA. There are 520 species in this family, assigned to 14 genera. Diseases associated with this family include: bright yellow mosaic, yellow mosaic, yellow mottle, leaf curling, stunting, streaks, reduced yields. They have single-stranded circular DNA genomes encoding genes that diverge in both directions from a virion strand origin of replication (i.e. geminivirus genomes are ambisense). According to the Baltimore classification they are considered class II viruses. It is the largest known family of single stranded DNA viruses. Mastrevirus and curtovirus Transmission (medicine), transmission is via various leafhopper species (e.g. maize streak virus and other African streak viruses are transmitted by ''Cicadulina mbila''), the only known topocuvirus species, ''Tomato pseudo-curly top virus'', is transmitted by the treehopper ''Micrutalis malleifera'', ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
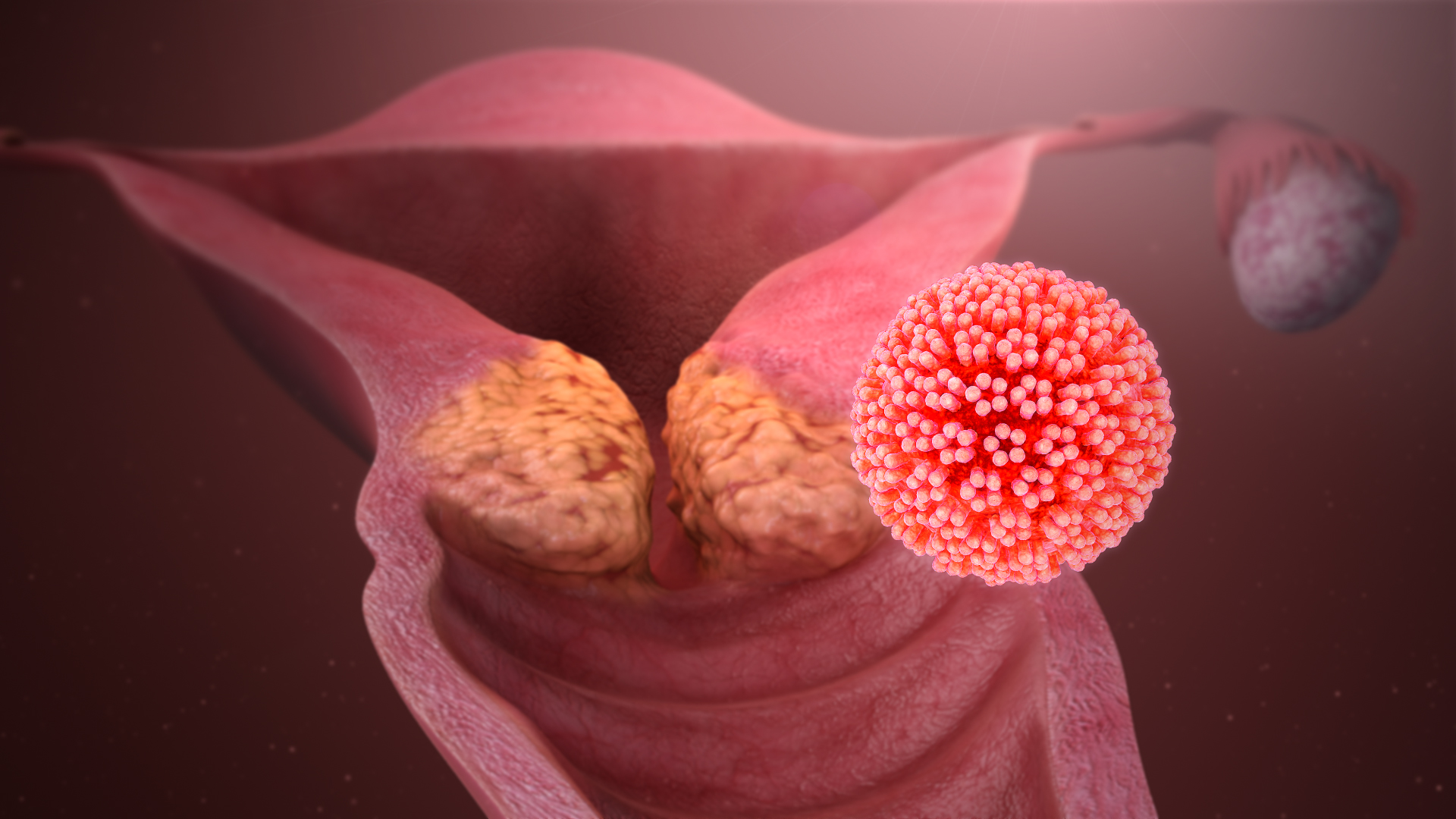
Human Papillomavirus Infection
Human papillomavirus infection (HPV infection) is caused by a DNA virus from the ''Papillomaviridae'' family. Many HPV infections cause no symptoms and 90% resolve spontaneously within two years. In some cases, an HPV infection persists and results in either warts or precancerous lesions. These lesions, depending on the site affected, increase the risk of cancer of the cervix, vulva, vagina, penis, anus, mouth, tonsils, or throat. Nearly all cervical cancer is due to HPV and two strains – HPV16 and HPV18 – account for 70% of cases. HPV16 is responsible for almost 90% of HPV-positive oropharyngeal cancers. Between 60% and 90% of the other cancers listed above are also linked to HPV. HPV6 and HPV11 are common causes of genital warts and laryngeal papillomatosis. An HPV infection is caused by ''human papillomavirus'', a DNA virus from the papillomavirus family. Over 170 types have been described. An individual can become infected with more than one type of HPV, and the disea ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Rolling Circle Replication Virology
Rolling is a type of motion that combines rotation (commonly, of an axially symmetric object) and translation of that object with respect to a surface (either one or the other moves), such that, if ideal conditions exist, the two are in contact with each other without sliding. Rolling where there is no sliding is referred to as ''pure rolling''. By definition, there is no sliding when there is a frame of reference in which all points of contact on the rolling object have the same velocity as their counterparts on the surface on which the object rolls; in particular, for a frame of reference in which the rolling plane is at rest (see animation), the instantaneous velocity of all the points of contact (e.g., a generating line segment of a cylinder) of the rolling object is zero. In practice, due to small deformations near the contact area, some sliding and energy dissipation occurs. Nevertheless, the resulting rolling resistance is much lower than sliding friction, and thus, roll ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribozymes
Ribozymes (ribonucleic acid enzymes) are RNA molecules that have the ability to catalyze specific biochemical reactions, including RNA splicing in gene expression, similar to the action of protein enzymes. The 1982 discovery of ribozymes demonstrated that RNA can be both genetic material (like DNA) and a biological catalyst (like protein enzymes), and contributed to the RNA world hypothesis, which suggests that RNA may have been important in the evolution of prebiotic self-replicating systems. The most common activities of natural or in vitro-evolved ribozymes are the cleavage or ligation of RNA and DNA and peptide bond formation. For example, the smallest ribozyme known (GUGGC-3') can aminoacylate a GCCU-3' sequence in the presence of PheAMP. Within the ribosome, ribozymes function as part of the large subunit ribosomal RNA to link amino acids during protein synthesis. They also participate in a variety of RNA processing reactions, including RNA splicing, viral replication, an ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Concatemers
A concatemer is a long continuous DNA molecule that contains multiple copies of the same DNA sequence linked in series. These polymeric molecules are usually copies of an entire genome linked end to end and separated by ''cos'' sites (a protein binding nucleotide sequence that occurs once in each copy of the genome). Concatemers are frequently the result of rolling circle replication, and may be seen in the late stage of bacterial infection by phages. As an example, if the genes in the phage DNA are arranged ABC, then in a concatemer the genes would be ABCABCABCABC and so on (assuming synthesis was initiated between genes C and A). They are further broken by ribozymes. During active infection, some species of viruses have been shown to replicate their genetic material via the formation of concatemers. In the case of ''human herpesvirus-6'', its entire genome is made over and over on a single strand. These long concatemers are subsequently cleaved between the pac-1 and pac-2 region ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Human Herpesvirus-6
Human herpesvirus 6 (HHV-6) is the common collective name for ''human betaherpesvirus 6A'' (HHV-6A) and ''human betaherpesvirus 6B'' (HHV-6B). These closely related viruses are two of the nine known human herpesviruses, herpesviruses that have humans as their primary host. HHV-6A and HHV-6B are DNA virus#Double-stranded DNA viruses, double-stranded DNA viruses within the ''Betaherpesvirinae'' subfamily and of the genus ''Roseolovirus''. HHV-6A and HHV-6B infect almost all of the human populations that have been tested. HHV-6A has been described as more neurovirulent, and as such is more frequently found in patients with neuroinflammatory diseases such as multiple sclerosis. HHV-6 (and HHV-7) levels in the brain are also elevated in people with Alzheimer's disease. HHV-6B primary infection is the cause of the common childhood illness exanthema subitum (also known as roseola infantum or sixth disease). It is passed on from child to child. It is uncommon for adults to contract this ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Virus
A DNA virus is a virus that has a genome made of deoxyribonucleic acid (DNA) that is replicated by a DNA polymerase. They can be divided between those that have two strands of DNA in their genome, called double-stranded DNA (dsDNA) viruses, and those that have one strand of DNA in their genome, called single-stranded DNA (ssDNA) viruses. dsDNA viruses primarily belong to two realms: ''Duplodnaviria'' and ''Varidnaviria'', and ssDNA viruses are almost exclusively assigned to the realm ''Monodnaviria'', which also includes some dsDNA viruses. Additionally, many DNA viruses are unassigned to higher taxa. Reverse transcribing viruses, which have a DNA genome that is replicated through an RNA intermediate by a reverse transcriptase, are classified into the kingdom '' Pararnavirae'' in the realm '' Riboviria''. DNA viruses are ubiquitous worldwide, especially in marine environments where they form an important part of marine ecosystems, and infect both prokaryotes and eukaryotes. The ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Okazaki Fragment
Okazaki fragments are short sequences of DNA nucleotides (approximately 150 to 200 base pairs long in eukaryotes) which are synthesized discontinuously and later linked together by the enzyme DNA ligase to create the lagging strand during DNA replication. They were discovered in the 1960s by the Japanese molecular biologists Reiji and Tsuneko Okazaki, along with the help of some of their colleagues. During DNA replication, the double helix is unwound and the complementary strands are separated by the enzyme DNA helicase, creating what is known as the DNA replication fork. Following this fork, DNA primase and DNA polymerase begin to act in order to create a new complementary strand. Because these enzymes can only work in the 5’ to 3’ direction, the two unwound template strands are replicated in different ways. One strand, the leading strand, undergoes a continuous replication process since its template strand has 3’ to 5’ directionality, allowing the polymerase assembl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |