|
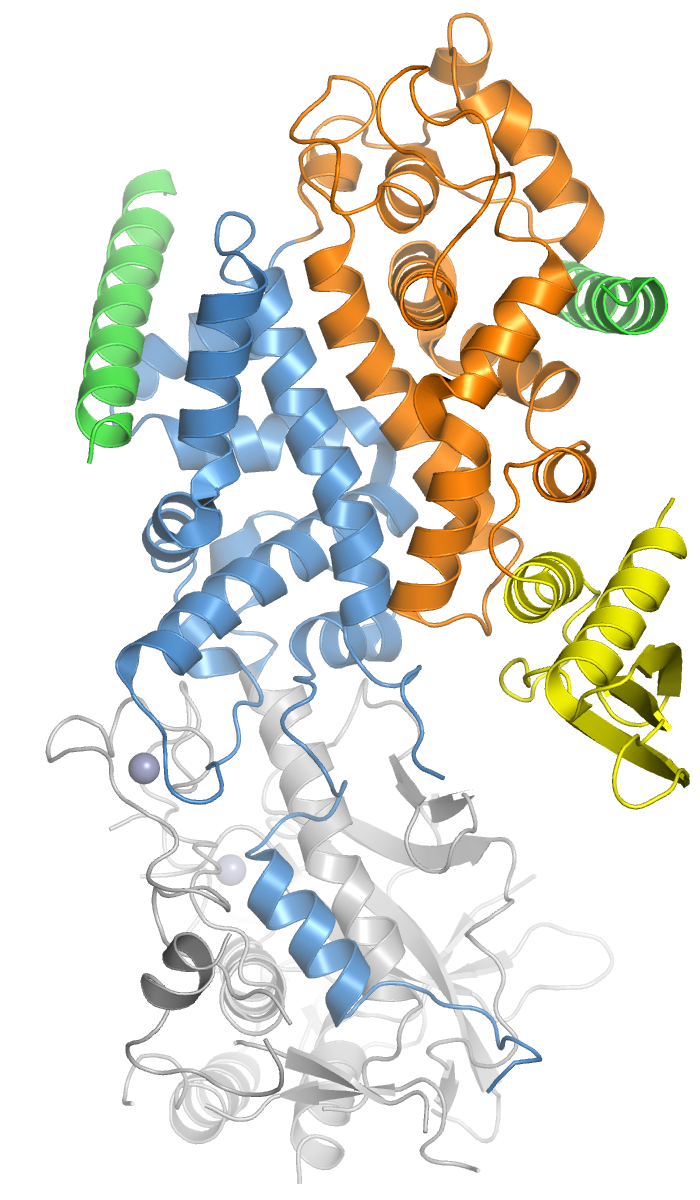
Ribonuclease III
Ribonuclease III (RNase III or RNase C)(BREND3.1.26.3 is a type of ribonuclease that recognizes dsRNA and cleaves it at specific targeted locations to transform them into mature RNAs. These enzymes are a group of endoribonucleases that are characterized by their ribonuclease domain, which is labelled the RNase III domain. They are ubiquitous compounds in the cell and play a major role in pathways such as RNA precursor synthesis, RNA Silencing, and the ''pnp'' autoregulatory mechanism. Types of RNase III The RNase III superfamily is divided into four known classes: 1, 2, 3, and 4. Each class is defined by its domain structure.Liang Y-H, Lavoie M, Comeau M-A, Elela SA, Ji X. Structure of a Eukaryotic RNase III Post-Cleavage Complex Reveals a Double- Ruler Mechanism for Substrate Selection. Molecular cell. 2014;54(3):431-444. doi:10.1016/j.molcel.2014.03.006. Class 1 RNase III *Class 1 RNase III enzymes have a homodimeric structure whose function is to cleave dsRNA into multip ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribonuclease
Ribonuclease (commonly abbreviated RNase) is a type of nuclease that catalyzes the degradation of RNA into smaller components. Ribonucleases can be divided into endoribonucleases and exoribonucleases, and comprise several sub-classes within the EC 2.7 (for the phosphorolytic enzymes) and 3.1 (for the hydrolytic enzymes) classes of enzymes. Function All organisms studied contain many RNases of two different classes, showing that RNA degradation is a very ancient and important process. As well as clearing of cellular RNA that is no longer required, RNases play key roles in the maturation of all RNA molecules, both messenger RNAs that carry genetic material for making proteins and non-coding RNAs that function in varied cellular processes. In addition, active RNA degradation systems are the first defense against RNA viruses and provide the underlying machinery for more advanced cellular immune strategies such as RNAi. Some cells also secrete copious quantities of non-specifi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribonucleases
Ribonuclease (commonly abbreviated RNase) is a type of nuclease that catalyzes the degradation of RNA into smaller components. Ribonucleases can be divided into endoribonucleases and exoribonucleases, and comprise several sub-classes within the EC 2.7 (for the phosphorolytic enzymes) and 3.1 (for the hydrolytic enzymes) classes of enzymes. Function All organisms studied contain many RNases of two different classes, showing that RNA degradation is a very ancient and important process. As well as clearing of cellular RNA that is no longer required, RNases play key roles in the maturation of all RNA molecules, both messenger RNAs that carry genetic material for making proteins and non-coding RNAs that function in varied cellular processes. In addition, active RNA degradation systems are the first defense against RNA viruses and provide the underlying machinery for more advanced cellular immune strategies such as RNAi. Some cells also secrete copious quantities of non-specific ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RncO
RncO is a bacterial non-coding RNA regulatory element found in the rnc leader sequence. The rnc operon is negatively auto-regulated by transcript stability. ''rnc'', the first gene in the operon codes for RNase III which cleaves the long rncO stem II leading to transcript degradation and a reduction in translation. Matsunaga ''et al.'' showed that RNase III cleavage can initiate rnc transcript decay independently of rnc gene translation. Further work has established that rncO structure and function is conserved in ''Salmonella typhimurium''. Structure Functionally the first 215 nucleotides of the rnc leader have been shown to be sufficient. Within this region three stem-loop Stem-loop intramolecular base pairing is a pattern that can occur in single-stranded RNA. The structure is also known as a hairpin or hairpin loop. It occurs when two regions of the same strand, usually complementary in nucleotide sequence wh ...s were identified. Stem-loop II is cleaved by RNase III, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DROSHA
Drosha is a Class 2 ribonuclease III enzyme that in humans is encoded by the ''DROSHA'' (formerly ''RNASEN'') gene. It is the primary nuclease that executes the initiation step of miRNA processing in the nucleus. It works closely with DGCR8 and in correlation with Dicer. It has been found significant in clinical knowledge for cancer prognosisSlack FJ, Weidhaas JB (December 2008). "MicroRNA in cancer prognosis". ''The New England Journal of Medicine.'' 359 (25): 2720-2. and HIV-1 replication.Swaminathan, G., Navas-Martín, S., & Martín-García, J. (2014). MicroRNAs and HIV-1 infection: antiviral activities and beyond. ''Journal of molecular biology'', ''426''(6), 1178-1197. History Human Drosha was cloned in 2000 when it was identified as a nuclear dsRNA ribonuclease involved in the processing of ribosomal RNA precursors. The other two human enzymes that participate in the processing and activity of miRNA are the Dicer and Argonaute proteins. Recently, proteins like Drosha h ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DICER1
Dicer, also known as endoribonuclease Dicer or helicase with RNase motif, is an enzyme that in humans is encoded by the gene. Being part of the RNase III family, Dicer cleaves double-stranded RNA (dsRNA) and pre-microRNA (pre-miRNA) into short double-stranded RNA fragments called small interfering RNA and microRNA, respectively. These fragments are approximately 20–25 base pairs long with a two-base overhang on the 3′-end. Dicer facilitates the activation of the RNA-induced silencing complex (RISC), which is essential for RNA interference. RISC has a catalytic component Argonaute, which is an endonuclease capable of degrading messenger RNA (mRNA). Discovery Dicer was given its name in 2001 by Stony Brook PhD student Emily Bernstein while conducting research in Gregory Hannon's lab at Cold Spring Harbor Laboratory. Bernstein sought to discover the enzyme responsible for generating small RNA fragments from double-stranded RNA. Dicer's ability to generate around 22 nuc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gregory Hannon
Gregory James Hannon One or more of the preceding sentences incorporates text from the royalsociety.org website where: (born 1964) is a professor of molecular cancer biology and director of the Cancer Research UK Cambridge Institute at the University of Cambridge. He is a Fellow of Trinity College, Cambridge while also serving as a director of cancer genomics at the New York Genome Center and an adjunct professor at Cold Spring Harbor Laboratory. Career and research Hannon is known for his contributions to small RNA biology, cancer biology, and mammalian genomics. He has a history in discovery of oncogenes, beginning with work that led to the identification of CDK inhibitors and their links to cancer. More recently, his work has focused on small RNA biology, which led to an understanding of the biochemical mechanisms and biological functions of RNA interference (RNAi). He has developed widely used tools and strategies for manipulation of gene expression in mammalian cells ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Emily Bernstein
Emily Bernstein is a professor at Mount Sinai School of Medicine known for her research on RNA interference, epigenetics, and cancer, especially melanoma. Education and career Bernstein received her B.S. from McGill University in 1998 and earned a Ph.D. from Stony Brook University in 2003. Following her Ph.D. she was a postdoctoral researcher at Rockefeller University where she worked with David Altis. In 2008 she moved to Mount Sinai School of Medicine where, as of 2022, she is a professor in the department of oncology and dermatology. Research Bernstein is known for her research on RNA interference, epigenetics, and cell development. Her early research examined the enzyme Dicer, its role in cell development in mice, and RNA interference. While a postdoctoral researcher she examined linkages between non-coding RNA and chromatin and DNA methylation. Subsequently, she has worked on histones, gene silencing, and tumor cell development. In 2022 her team discovered alterations t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Interference
RNA interference (RNAi) is a biological process in which RNA molecules are involved in sequence-specific suppression of gene expression by double-stranded RNA, through translational or transcriptional repression. Historically, RNAi was known by other names, including ''co-suppression'', ''post-transcriptional gene silencing'' (PTGS), and ''quelling''. The detailed study of each of these seemingly different processes elucidated that the identity of these phenomena were all actually RNAi. Andrew Fire and Craig C. Mello shared the 2006 Nobel Prize in Physiology or Medicine for their work on RNAi in the nematode worm '' Caenorhabditis elegans'', which they published in 1998. Since the discovery of RNAi and its regulatory potentials, it has become evident that RNAi has immense potential in suppression of desired genes. RNAi is now known as precise, efficient, stable and better than antisense therapy for gene suppression. Antisense RNA produced intracellularly by an expression vect ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dicer
Dicer, also known as endoribonuclease Dicer or helicase with RNase motif, is an enzyme that in humans is encoded by the gene. Being part of the RNase III family, Dicer cleaves double-stranded RNA (dsRNA) and pre-microRNA (pre-miRNA) into short double-stranded RNA fragments called small interfering RNA and microRNA, respectively. These fragments are approximately 20–25 base pairs long with a two-base overhang on the 3′-end. Dicer facilitates the activation of the RNA-induced silencing complex (RISC), which is essential for RNA interference. RISC has a catalytic component Argonaute, which is an endonuclease capable of degrading messenger RNA (mRNA). Discovery Dicer was given its name in 2001 by Stony Brook PhD student Emily Bernstein while conducting research in Gregory Hannon's lab at Cold Spring Harbor Laboratory. Bernstein sought to discover the enzyme responsible for generating small RNA fragments from double-stranded RNA. Dicer's ability to generate around 22 n ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MicroRNA
MicroRNA (miRNA) are small, single-stranded, non-coding RNA molecules containing 21 to 23 nucleotides. Found in plants, animals and some viruses, miRNAs are involved in RNA silencing and post-transcriptional regulation of gene expression. miRNAs base-pair to complementary sequences in mRNA molecules, then gene silence said mRNA molecules by one or more of the following processes: (1) cleavage of mRNA strand into two pieces, (2) destabilization of mRNA by shortening its poly(A) tail, or (3) translation of mRNA into proteins. This last method of gene silencing is the least efficient of the three, and requires the aid of ribosomes. miRNAs resemble the small interfering RNAs (siRNAs) of the RNA interference (RNAi) pathway, except miRNAs derive from regions of RNA transcripts that fold back on themselves to form short hairpins, whereas siRNAs derive from longer regions of double-stranded RNA. The human genome may encode over 1900 miRNAs, although more recent analysis sugges ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Drosha
Drosha is a Class 2 ribonuclease III enzyme that in humans is encoded by the ''DROSHA'' (formerly ''RNASEN'') gene. It is the primary nuclease that executes the initiation step of miRNA processing in the nucleus. It works closely with DGCR8 and in correlation with Dicer. It has been found significant in clinical knowledge for cancer prognosisSlack FJ, Weidhaas JB (December 2008). "MicroRNA in cancer prognosis". ''The New England Journal of Medicine.'' 359 (25): 2720-2. and HIV-1 replication.Swaminathan, G., Navas-Martín, S., & Martín-García, J. (2014). MicroRNAs and HIV-1 infection: antiviral activities and beyond. ''Journal of molecular biology'', ''426''(6), 1178-1197. History Human Drosha was cloned in 2000 when it was identified as a nuclear dsRNA ribonuclease involved in the processing of ribosomal RNA precursors. The other two human enzymes that participate in the processing and activity of miRNA are the Dicer and Argonaute proteins. Recently, proteins like Drosha h ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |