|
Quantitative Proteomics
Quantitative proteomics is an analytical chemistry technique for determining the amount of proteins in a sample. The methods for protein identification are identical to those used in general (i.e. qualitative) proteomics, but include quantification as an additional dimension. Rather than just providing lists of proteins identified in a certain sample, quantitative proteomics yields information about the physiological differences between two biological samples. For example, this approach can be used to compare samples from healthy and diseased patients. Quantitative proteomics is mainly performed by two-dimensional gel electrophoresis (2-DE) or mass spectrometry (MS). However, a recent developed method of quantitative dot blot (QDB) analysis is able to measure both the absolute and relative quantity of an individual proteins in the sample in high throughput format, thus open a new direction for proteomic research. In contrast to 2-DE, which requires MS for the downstream protein id ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Quantitative Mass Spectrometry
Quantitative may refer to: * Quantitative research, scientific investigation of quantitative properties * Quantitative analysis (other) * Quantitative verse, a metrical system in poetry * Statistics, also known as quantitative analysis * Numerical data, also known as quantitative data * Quantification (science) In mathematics and empirical science, quantification (or quantitation) is the act of counting and measuring that maps human sense observations and experiences into quantities. Quantification in this sense is fundamental to the scientific method ... See also * Qualitative {{disambig ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Isotopologues
In chemistry, isotopologues are molecules that differ only in their isotopic composition. They have the same chemical formula and bonding arrangement of atoms, but at least one atom has a different number of neutrons than the parent. An example is water, whose hydrogen-related isotopologues are: "light water" (HOH or ), " semi-heavy water" with the deuterium isotope in equal proportion to protium (HDO or ), " heavy water" with two deuterium isotopes of hydrogen per molecule ( or ), and "super-heavy water" or tritiated water ( or , as well as and , where some or all of the hydrogen atoms are replaced with the radioactive tritium isotope). Oxygen-related isotopologues of water include the commonly available form of heavy-oxygen water () and the more difficult to separate version with the isotope. Both elements may be replaced by isotopes, for example in the doubly labeled water isotopologue . All taken together, there are 9 different stable water isotopologues, and 9 radioactiv ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Label-free Quantification
Label-free quantification is a method in mass spectrometry that aims to determine the relative amount of proteins in two or more biological samples. Unlike other methods for protein quantification, label-free quantification does not use a stable isotope containing compound to chemically bind to and thus label the protein. Implementation Label-free quantification may be based on precursor signal intensity or on spectral counting. The first method is useful when applied to high precision mass spectra, such as those obtained using the new generation of time-of-flight (ToF), fourier transform ion cyclotron resonance (FTICR), or Orbitrap mass analyzers. The high-resolution power facilitates the extraction of peptide signals on the MS1 level and thus uncouples the quantification from the identification process. In contrast, spectral counting simply counts the number of spectra identified for a given peptide in different biological samples and then integrates the results for all measur ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MALDI
In mass spectrometry, matrix-assisted laser desorption/ionization (MALDI) is an ionization technique that uses a laser energy absorbing matrix to create ions from large molecules with minimal fragmentation. It has been applied to the analysis of biomolecules ( biopolymers such as DNA, proteins, peptides and carbohydrates) and various organic molecules (such as polymers, dendrimers and other macromolecules), which tend to be fragile and fragment when ionized by more conventional ionization methods. It is similar in character to electrospray ionization (ESI) in that both techniques are relatively soft (low fragmentation) ways of obtaining ions of large molecules in the gas phase, though MALDI typically produces far fewer multi-charged ions. MALDI methodology is a three-step process. First, the sample is mixed with a suitable matrix material and applied to a metal plate. Second, a pulsed laser irradiates the sample, triggering ablation and desorption of the sample and matrix materi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ICP-MS
Inductively coupled plasma mass spectrometry (ICP-MS) is a type of mass spectrometry that uses an inductively coupled plasma to Ionization, ionize the sample. It atomizes the sample and creates atomic and small polyatomic ions, which are then detected. It is known and used for its ability to detect metals and several non-metals in liquid samples at very low concentrations. It can detect different isotopes of the same element, which makes it a versatile tool in isotopic labeling. Compared to atomic absorption spectroscopy, ICP-MS has greater speed, precision, and sensitivity. However, compared with other types of mass spectrometry, such as thermal ionization mass spectrometry (TIMS) and glow discharge mass spectrometry (GD-MS), ICP-MS introduces many interfering species: argon from the plasma, component gases of air that leak through the cone orifices, and contamination from glassware and the cones. Components Inductively coupled plasma An inductively coupled plasma is a Plasma ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
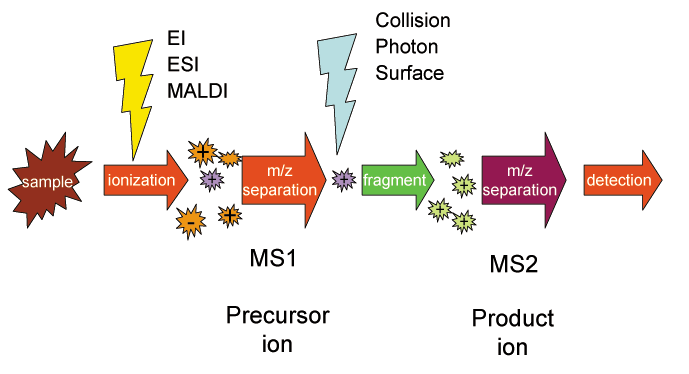
Selected Reaction Monitoring
Selected reaction monitoring (SRM), also called Multiple reaction monitoring, (MRM), is a method used in tandem mass spectrometry in which an ion of a particular mass is selected in the first stage of a tandem mass spectrometer and an ion product of a fragmentation reaction of the precursor ions is selected in the second mass spectrometer stage for detection. Variants A general case of SRM can be represented by :ABCD^+ \to AB + CD^+ where the precursor ion ABCD+ is selected by the first stage of mass spectrometry (MS1), dissociates into molecule AB and product ion CD+, and the latter is selected by the second stage of mass spectrometry (MS2) and detected. The precursor and product ion pair is called a SRM "transition." Consecutive reaction monitoring (CRM) is the serial application of three or more stages of mass spectrometry to SRM, represented in a simple case by :ABCD^+ \to AB + CD^+ \to C + D^+ where ABCD+ is selected by MS1, dissociates into molecule AB and ion CD+. Th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Terminal Amine Isotopic Labeling Of Substrates
Terminal amine isotopic labeling of substrates (TAILS) is a method in quantitative proteomics that identifies the protein content of samples based on N-terminal fragments of each protein (N-terminal peptides) and detects differences in protein abundance among samples. Like other methods based on N-terminal peptides, this assay uses trypsin to break proteins into fragments and separates the N-terminal peptides (the fragments containing the N-termini of the original proteins) from the other fragments (internal tryptic peptides). TAILS isolates the N-terminal peptides by identifying and removing the internal tryptic peptides. This negative selection allows the TAILS method to detect all N-termini in the given samples. Alternative methods that rely on the free amino group of the N-terminus to identify the N-terminal peptides cannot detect some N-termini because they are "naturally blocked" (i.e. the natural protein does not have a free amino group). The TAILS method has a number of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SILAC
Stable Isotope Labeling by/with Amino acids in Cell culture (SILAC) is a technique based on mass spectrometry that detects differences in protein abundance among samples using non-radioactive isotopic labeling. It is a popular method for quantitative proteomics. Procedure Two populations of cells are cultivated in cell culture. One of the cell populations is fed with growth medium containing normal amino acids. In contrast, the second population is fed with growth medium containing amino acids labeled with stable (non-radioactive) heavy isotopes. For example, the medium can contain arginine labeled with six carbon-13 atoms (13C) instead of the normal carbon-12 (12C). When the cells are growing in this medium, they incorporate the heavy arginine into all of their proteins. Thereafter, all peptides containing a single arginine are 6 Da heavier than their normal counterparts. Alternatively, uniform labeling with 13C or 15N can be used. Proteins from both cell populations are combined ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
N-terminal Labelling
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the amine group is bonded to the carboxylic group of another amino acid, making it a chain. That leaves a free carboxylic group at one end of the peptide, called the C-terminus, and a free amine group on the other end called the N-terminus. By convention, peptide sequences are written N-terminus to C-terminus, left to right (in LTR writing systems). This correlates the translation direction to the text direction, because when a protein is translated from messenger RNA, it is created from the N-terminus to the C-terminus, as amino acids are added to the carboxyl end of the protein. Chemistry Each amino acid has an amine group and a carboxylic group. Amino acids link to one another by peptide bonds which form through a dehydration reaction t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MeCAT
Metal-coded affinity tag is a method used for quantitative proteomics by mass spectrometry that uses a metal chelate complex 1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetate (DOTA) coupled to different lanthanide ions. The metal complexes attach to the cysteine residues of proteins in a sample. Proteomic analysis For bottom-up proteomics, the proteins can be separated by two-dimensional gel electrophoresis and analyzed by matrix-assisted laser desorption/ionization (MALDI) or electrospray ionization mass spectrometry for relative quantification or by inductively coupled plasma mass spectrometry for absolute quantification. For top-down proteomics Top-down proteomics is a method of protein identification that either uses an ion trapping mass spectrometer to store an isolated protein ion for mass measurement and tandem mass spectrometry (MS/MS) analysis or other protein purification methods ..., the undigested labeled proteins are analyzed. See also * Mass cytometry Refere ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Label-free Quantification
Label-free quantification is a method in mass spectrometry that aims to determine the relative amount of proteins in two or more biological samples. Unlike other methods for protein quantification, label-free quantification does not use a stable isotope containing compound to chemically bind to and thus label the protein. Implementation Label-free quantification may be based on precursor signal intensity or on spectral counting. The first method is useful when applied to high precision mass spectra, such as those obtained using the new generation of time-of-flight (ToF), fourier transform ion cyclotron resonance (FTICR), or Orbitrap mass analyzers. The high-resolution power facilitates the extraction of peptide signals on the MS1 level and thus uncouples the quantification from the identification process. In contrast, spectral counting simply counts the number of spectra identified for a given peptide in different biological samples and then integrates the results for all measur ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Isobaric Tags For Relative And Absolute Quantification
Isobaric tags for relative and absolute quantitation (iTRAQ) is an isobaric labeling method used in quantitative proteomics by tandem mass spectrometry to determine the amount of proteins from different sources in a single experiment. It uses stable isotope labeled molecules that can be covalent bonded to the N-terminus and side chain amines of proteins. Procedure The ITRAQ method is based on the covalent labeling of the N-terminus and side chain amines of peptides from protein digestions with tags of varying mass. There are currently two mainly used reagents: 4-plex and 8-plex, which can be used to label all peptides from different samples/treatments. These samples are then pooled and usually fractionated by liquid chromatography In chemical analysis, chromatography is a laboratory technique for the separation of a mixture into its components. The mixture is dissolved in a fluid solvent (gas or liquid) called the ''mobile phase'', which carries it through a system ( ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |