|
Peptide Plane Flipping
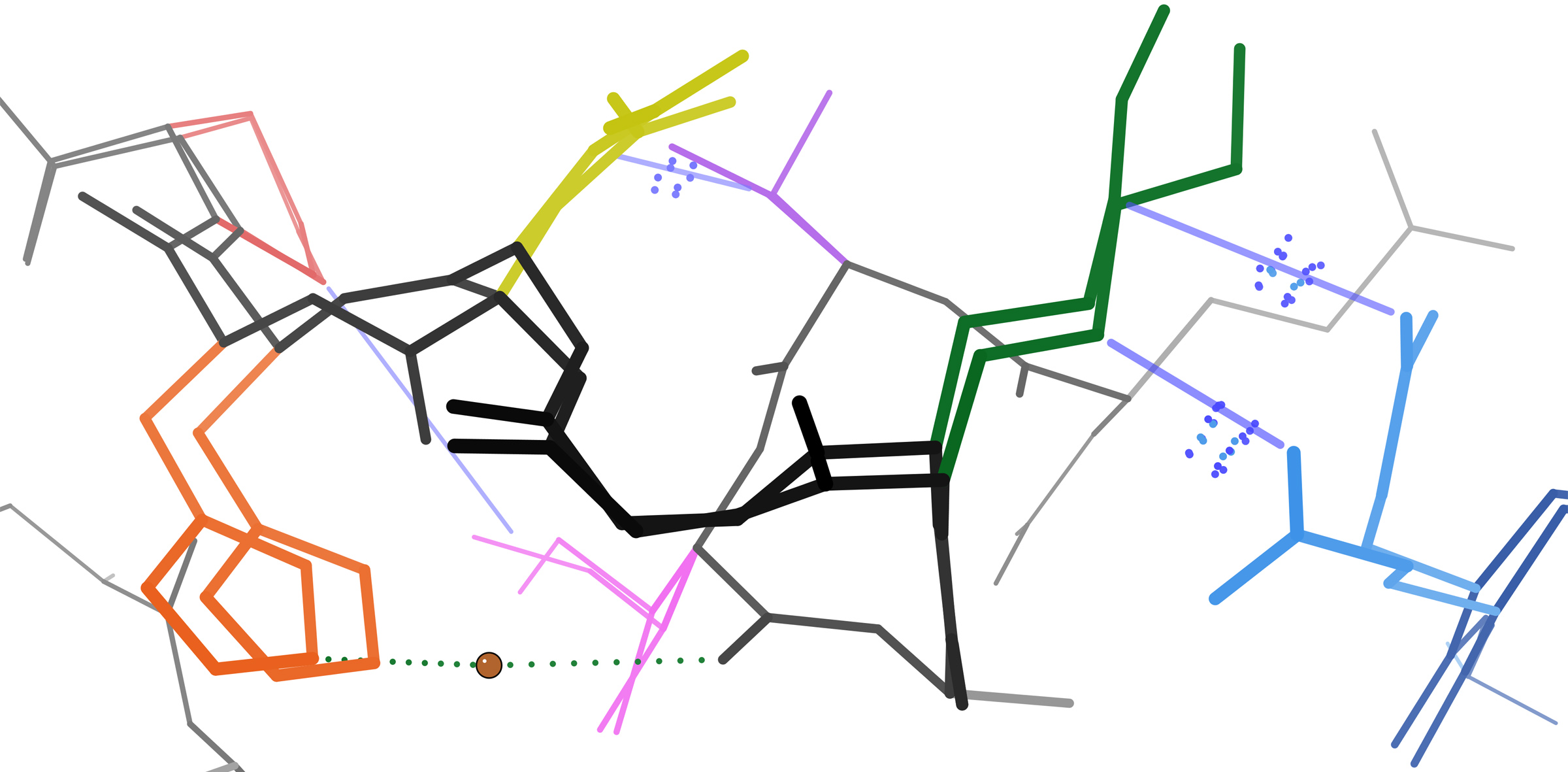
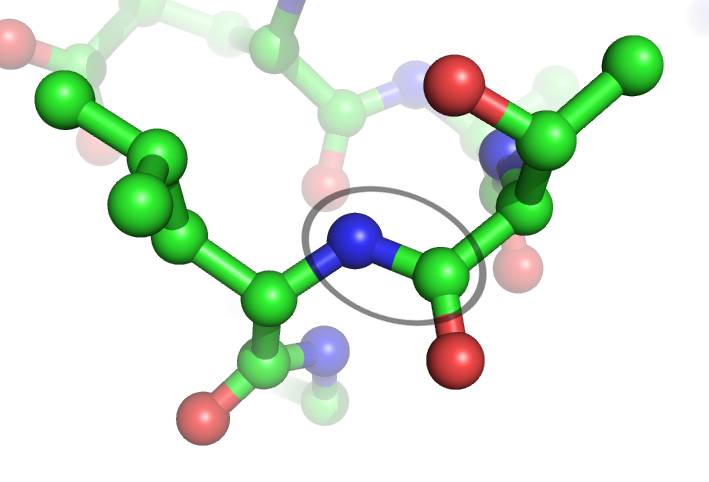
Peptide plane flipping is a type of conformational change that can occur in proteins by which the dihedral angles of adjacent amino acids undergo large-scale rotations with little displacement of the side chains. The plane flip is defined as a rotation of the dihedral angles φ,ψ at amino acids ''i'' and ''i+1'' such that the resulting angles remain in structurally stable regions of Ramachandran space. The key requirement is that the ''sum'' of the ψ''i'' angle of residue ''i'' and the φ''i+1'' angle of residue ''i+1'' remain roughly constant; in effect, the flip is a crankshaft move about the axis defined by the Cα-C¹ and N-Cα bond vectors of the peptide group, which are roughly parallel. As an example, the type I and type II beta turns differ by a simple flip of the central peptide group of the turn. In protein dynamics The significance of peptide plane flips in the dynamics of the native state has been inferred in some proteins by comparing crystal structures of the sa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Conformational Change
In biochemistry, a conformational change is a change in the shape of a macromolecule, often induced by environmental factors. A macromolecule is usually flexible and dynamic. Its shape can change in response to changes in its environment or other factors; each possible shape is called a conformation, and a transition between them is called a ''conformational change''. Factors that may induce such changes include temperature, pH, voltage, light in chromophores, concentration of ions, phosphorylation, or the binding of a ligand. Transitions between these states occur on a variety of length scales (tenths of Å to nm) and time scales (ns to s), and have been linked to functionally relevant phenomena such as allosteric signaling and enzyme catalysis. Laboratory analysis Many biophysical techniques such as crystallography, NMR, electron paramagnetic resonance (EPR) using spin label techniques, circular dichroism (CD), hydrogen exchange, and FRET can be used to study macrom ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Native State
In biochemistry, the native state of a protein or nucleic acid is its properly folded and/or assembled form, which is operative and functional. The native state of a biomolecule may possess all four levels of biomolecular structure, with the secondary through quaternary structure being formed from weak interactions along the covalently-bonded backbone. This is in contrast to the denatured state, in which these weak interactions are disrupted, leading to the loss of these forms of structure and retaining only the biomolecule's primary structure. Biochemistry Proteins While all protein molecules begin as simple unbranched chains of amino acids, once completed they assume highly specific three-dimensional shapes. That ultimate shape, known as tertiary structure, is the folded shape that possesses a minimum of free energy. It is a protein's tertiary, folded structure that makes it capable of performing its biological function. In fact, shape changes in proteins are the primary ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Folding
Protein folding is the physical process by which a protein chain is translated to its native three-dimensional structure, typically a "folded" conformation by which the protein becomes biologically functional. Via an expeditious and reproducible process, a polypeptide folds into its characteristic three-dimensional structure from a random coil. Each protein exists first as an unfolded polypeptide or random coil after being translated from a sequence of mRNA to a linear chain of amino acids. At this stage the polypeptide lacks any stable (long-lasting) three-dimensional structure (the left hand side of the first figure). As the polypeptide chain is being synthesized by a ribosome, the linear chain begins to fold into its three-dimensional structure. Folding of many proteins begins even during translation of the polypeptide chain. Amino acids interact with each other to produce a well-defined three-dimensional structure, the folded protein (the right hand side of the figure), ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alpha Sheet
Alpha sheet (also known as alpha pleated sheet or polar pleated sheet) is an atypical secondary structure in proteins, first proposed by Linus Pauling and Robert Corey in 1951.Pauling, L. & Corey, R. B. (1951). The pleated sheet, a new layer configuration of polypeptide chains. ''Proc. Natl. Acad. Sci. USA'' 37, 251–6. Pauling, L. & Corey, R. B. (1951). The structure of feather rachis keratin. ''Proc. Natl. Acad. Sci. USA'' 37, 256–261. Pauling, L. & Corey, R. B. (1951). Configurations of Polypeptide Chains With Favored Orientations Around Single Bonds: Two New Pleated Sheets. ''Proc. Natl. Acad. Sci. USA'' 37, 729–740. The hydrogen bonding pattern in an alpha sheet is similar to that of a beta sheet, but the orientation of the carbonyl and amino groups in the peptide bond units is distinctive; in a single strand, all the carbonyl groups are oriented in the same direction on one side of the pleat, and all the amino groups are oriented in the same direction on the opposite s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Beta Sheet
The beta sheet, (β-sheet) (also β-pleated sheet) is a common motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally by at least two or three backbone hydrogen bonds, forming a generally twisted, pleated sheet. A β-strand is a stretch of polypeptide chain typically 3 to 10 amino acids long with backbone in an extended conformation. The supramolecular association of β-sheets has been implicated in the formation of the fibrils and protein aggregates observed in amyloidosis, notably Alzheimer's disease. History The first β-sheet structure was proposed by William Astbury in the 1930s. He proposed the idea of hydrogen bonding between the peptide bonds of parallel or antiparallel extended β-strands. However, Astbury did not have the necessary data on the bond geometry of the amino acids in order to build accurate models, especially since he did not then know that the peptide bond was planar. A refined versi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Amyloid
Amyloids are aggregates of proteins characterised by a Fibril, fibrillar morphology of 7–13 Nanometer, nm in diameter, a beta sheet (β-sheet) Secondary structure of proteins, secondary structure (known as cross-β) and ability to be Staining, stained by particular dyes, such as Congo red. In the human body, amyloids have been linked to the development of various diseases. Pathogenic amyloids form when previously healthy proteins lose their normal Protein structure, structure and physiology, physiological functions (Protein misfolding, misfolding) and form fibrous deposits in amyloid plaques around cells which can disrupt the healthy function of tissues and organs. Such amyloids have been associated with (but not necessarily as the cause of) more than 50 human diseases, known as amyloidosis, and may play a role in some neurodegenerative diseases. Some of these diseases are mainly sporadic and only a few cases are Genetic disorder, familial. Others are only Genetic disorder, fam ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
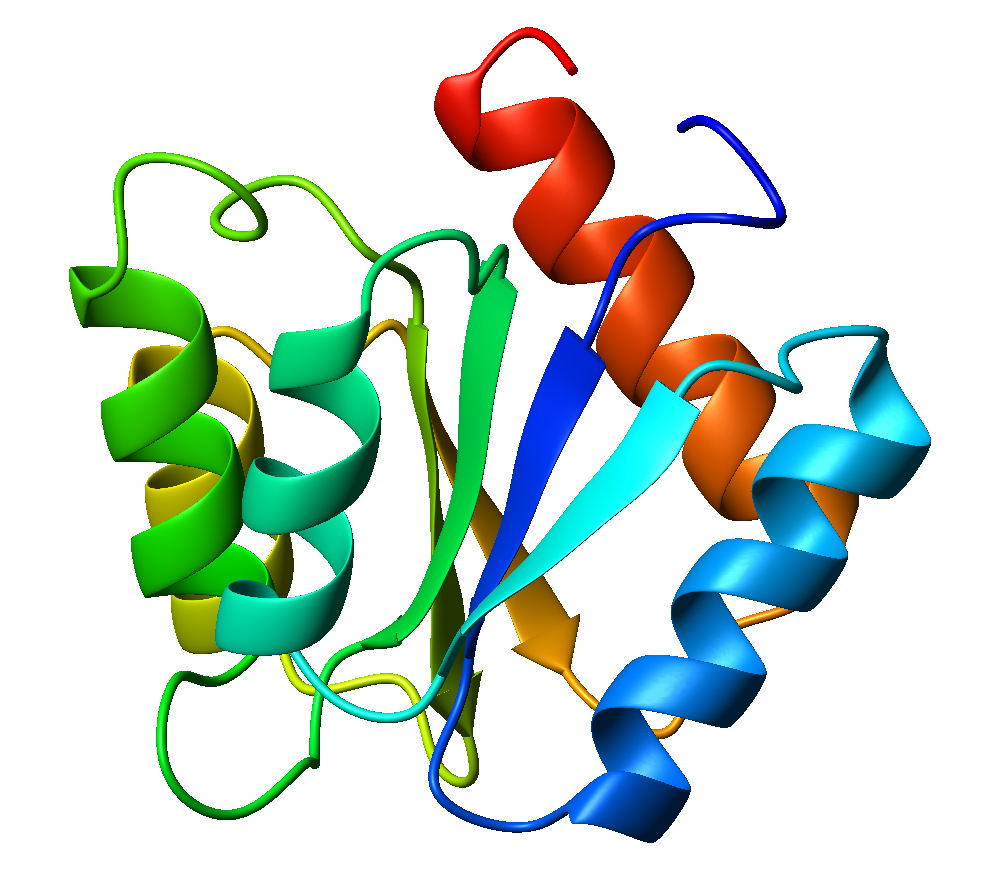
Flavodoxin
Flavodoxins (Fld) are small, soluble electron-transfer proteins. Flavodoxins contains flavin mononucleotide as prosthetic group. The structure of flavodoxin is characterized by a five-stranded parallel beta sheet, surrounded by five alpha helices. They have been isolated from prokaryotes, cyanobacteria, and some eukaryotic algae. Background Originally found in cyanobacteria and clostridia, flavodoxins were discovered over 50 years ago. These proteins evolved from an anaerobic environment, due to selective pressures. Ferredoxin, another redox protein, was the only protein able to be used in this manner. However, when oxygen became present in the environment, iron became limited. Ferredoxin is iron-dependant as well as oxidant-sensitive. Under these limited iron conditions, ferredoxin was no longer preferred. Flavodoxin on the other hand is the opposite of these traits, as it is oxidant-resistant and has iron-free isofunctional counterparts. Therefore, for some time flavodoxin was t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
X-ray Crystallography
X-ray crystallography is the experimental science determining the atomic and molecular structure of a crystal, in which the crystalline structure causes a beam of incident X-rays to diffract into many specific directions. By measuring the angles and intensities of these diffracted beams, a crystallographer can produce a three-dimensional picture of the density of electrons within the crystal. From this electron density, the mean positions of the atoms in the crystal can be determined, as well as their chemical bonds, their crystallographic disorder, and various other information. Since many materials can form crystals—such as salts, metals, minerals, semiconductors, as well as various inorganic, organic, and biological molecules—X-ray crystallography has been fundamental in the development of many scientific fields. In its first decades of use, this method determined the size of atoms, the lengths and types of chemical bonds, and the atomic-scale differences among various mat ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Dynamics
Proteins are generally thought to adopt unique structures determined by their amino acid sequences. However, proteins are not strictly static objects, but rather populate ensembles of (sometimes similar) conformations. Transitions between these states occur on a variety of length scales (tenths of Å to nm) and time scales (ns to s), and have been linked to functionally relevant phenomena such as allosteric signaling and enzyme catalysis. The study of protein dynamics is most directly concerned with the transitions between these states, but can also involve the nature and equilibrium populations of the states themselves. These two perspectives—kinetics and thermodynamics, respectively—can be conceptually synthesized in an "energy landscape" paradigm: highly populated states and the kinetics of transitions between them can be described by the depths of energy wells and the heights of energy barriers, respectively. Local flexibility: atoms and residues Portions of protein s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid residue ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Peptide Bond
In organic chemistry, a peptide bond is an amide type of covalent chemical bond linking two consecutive alpha-amino acids from C1 (carbon number one) of one alpha-amino acid and N2 (nitrogen number two) of another, along a peptide or protein chain. It can also be called a eupeptide bond to distinguish it from an isopeptide bond, which is another type of amide bond between two amino acids. Synthesis When two amino acids form a ''dipeptide'' through a ''peptide bond'', it is a type of condensation reaction. In this kind of condensation, two amino acids approach each other, with the non-side chain (C1) carboxylic acid moiety of one coming near the non-side chain (N2) amino moiety of the other. One loses a hydrogen and oxygen from its carboxyl group (COOH) and the other loses a hydrogen from its amino group (NH2). This reaction produces a molecule of water (H2O) and two amino acids joined by a peptide bond (−CO−NH−). The two joined amino acids are called a dipeptide. The am ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Turn (biochemistry)
A turn is an element of secondary structure in proteins where the polypeptide chain reverses its overall direction. Definition According to one definition, a turn is a structural motif where the Cα atoms of two residues separated by a few (usually 1 to 5) peptide bonds are close (less than ). The proximity of the terminal Cα atoms often correlates with formation of an inter main chain hydrogen bond between the corresponding residues. Such hydrogen bonding is the basis for the original, perhaps better known, turn definition. In many cases, but not all, the hydrogen-bonding and Cα-distance definitions are equivalent. Types of turns Turns are classified according to the separation between the two end residues: * In an α-turn the end residues are separated by ''four'' peptide bonds (''i'' → ''i'' ± 4). * In a β-turn (the most common form), by ''three'' bonds (''i'' → ''i'' ± 3). * In a γ-turn, by ''two'' bonds (''i'' → ''i'' ± 2). ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |