|
Polytomy
An internal node of a phylogenetic tree is described as a polytomy or multifurcation if (i) it is in a rooted tree and is linked to three or more child subtrees or (ii) it is in an unrooted tree and is attached to four or more branches. A tree that contains any multifurcations can be described as a multifurcating tree. Soft polytomies vs. hard polytomies Two types of polytomies are recognized, soft and hard polytomies. Soft polytomies are the result of insufficient phylogenetic information: though the lineages diverged at different times – meaning that some of these lineages are closer relatives than others, and the available data does not allow recognition of this. Most polytomies are soft, meaning that they would be resolved into a typical tree of dichotomies if better data were available. In contrast, a hard polytomy represents a true divergence event of three or more lineages. Applications Interpretations for a polytomy depend on the individuals that are represented ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Generation
A generation is all of the people born and living at about the same time, regarded collectively. It also is "the average period, generally considered to be about 20–30 years, during which children are born and grow up, become adults, and begin to have children." In kinship, ''generation'' is a structural term, designating the parent–child relationship. In biology, ''generation'' also means biogenesis, reproduction, and procreation. ''Generation'' is also a synonym for ''birth/age cohort'' in demographics, marketing, and social science, where it means "people within a delineated population who experience the same significant events within a given period of time." The term ''generation'' in this sense, also known as '' social generations'', is widely used in popular culture and is a basis of sociological analysis. Serious analysis of generations began in the nineteenth century, emerging from an increasing awareness of the possibility of permanent social change and the i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phylogenetics
In biology, phylogenetics () is the study of the evolutionary history of life using observable characteristics of organisms (or genes), which is known as phylogenetic inference. It infers the relationship among organisms based on empirical data and observed heritable traits of DNA sequences, protein amino acid sequences, and morphology. The results are a phylogenetic tree—a diagram depicting the hypothetical relationships among the organisms, reflecting their inferred evolutionary history. The tips of a phylogenetic tree represent the observed entities, which can be living taxa or fossils. A phylogenetic diagram can be rooted or unrooted. A rooted tree diagram indicates the hypothetical common ancestor of the taxa represented on the tree. An unrooted tree diagram (a network) makes no assumption about directionality of character state transformation, and does not show the origin or "root" of the taxa in question. In addition to their use for inferring phylogenetic pa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phylogenetic Comparative Methods
Phylogenetic comparative methods (PCMs) use information on the historical relationships of lineages (phylogenies) to test evolutionary hypotheses. The comparative method has a long history in evolutionary biology; indeed, Charles Darwin used differences and similarities between species as a major source of evidence in ''The Origin of Species''. However, the fact that closely related lineages share many traits and trait combinations as a result of the process of descent with modification means that lineages are not independent. This realization inspired the development of explicitly phylogenetic comparative methods. Initially, these methods were primarily developed to control for phylogenetic history when testing for adaptation; however, in recent years the use of the term has broadened to include any use of phylogenies in statistical tests. Although most studies that employ PCMs focus on extant organisms, many methods can also be applied to extinct taxa and can incorporate informati ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Computational Phylogenetics
Computational phylogenetics, phylogeny inference, or phylogenetic inference focuses on computational and optimization algorithms, Heuristic (computer science), heuristics, and approaches involved in Phylogenetics, phylogenetic analyses. The goal is to find a phylogenetic tree representing optimal evolutionary ancestry between a set of genes, species, or taxa. Maximum likelihood estimation, Maximum likelihood, Maximum parsimony (phylogenetics), parsimony, Bayesian inference in phylogeny, Bayesian, and minimum evolution are typical optimality criteria used to assess how well a phylogenetic tree topology describes the sequence data. Nearest Neighbour Interchange (NNI), Subtree Prune and Regraft (SPR), and Tree Bisection and Reconnection (TBR), known as tree rearrangements, are deterministic algorithms to search for optimal or the best phylogenetic tree. The space and the landscape of searching for the optimal phylogenetic tree is known as phylogeny search space. Maximum Likelihood (al ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cladistics
Cladistics ( ; from Ancient Greek 'branch') is an approach to Taxonomy (biology), biological classification in which organisms are categorized in groups ("clades") based on hypotheses of most recent common ancestry. The evidence for hypothesized relationships is typically shared derived (phylogenetics), derived characteristics (synapomorphies) that are not present in more distant groups and ancestors. However, from an empirical perspective, common ancestors are inferences based on a cladistic hypothesis of relationships of taxa whose Phenotypic trait, character states can be observed. Theoretically, a last common ancestor and all its descendants constitute a (minimal) clade. Importantly, all descendants stay in their overarching ancestral clade. For example, if the terms ''worms'' or ''fishes'' were used within a ''strict'' cladistic framework, these terms would include humans. Many of these terms are normally used Paraphyly, paraphyletically, outside of cladistics, e.g. as a 'E ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Clock
The molecular clock is a figurative term for a technique that uses the mutation rate of biomolecules to deduce the time in prehistory when two or more life forms diverged. The biomolecular data used for such calculations are usually nucleotide sequences for DNA, RNA, or amino acid sequences for proteins. Early discovery and genetic equidistance The notion of the existence of a so-called "molecular clock" was first attributed to Émile Zuckerkandl and Linus Pauling who, in 1962, noticed that the number of amino acid differences in hemoglobin between different lineages changes roughly linearly with time, as estimated from fossil evidence. They generalized this observation to assert that the rate of evolutionary change of any specified protein was approximately constant over time and over different lineages (known as the molecular clock hypothesis). The genetic equidistance phenomenon was first noted in 1963 by Emanuel Margoliash, who wrote: "It appears that the number ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genetic Drift
Genetic drift, also known as random genetic drift, allelic drift or the Wright effect, is the change in the Allele frequency, frequency of an existing gene variant (allele) in a population due to random chance. Genetic drift may cause gene variants to disappear completely and thereby reduce genetic variation. It can also cause initially rare alleles to become much more frequent and even fixed. When few copies of an allele exist, the effect of genetic drift is more notable, and when many copies exist, the effect is less notable (due to the law of large numbers). In the middle of the 20th century, vigorous debates occurred over the relative importance of natural selection versus neutral processes, including genetic drift. Ronald Fisher, who explained natural selection using Mendelian inheritance, Mendelian genetics, held the view that genetic drift plays at most a minor role in evolution, and this remained the dominant view for several decades. In 1968, population geneticist Mot ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Founder Effect
In population genetics, the founder effect is the loss of genetic variation that occurs when a new population is established by a very small number of individuals from a larger population. It was first fully outlined by Ernst Mayr in 1942, using existing theoretical work by those such as Sewall Wright. As a result of the loss of genetic variation, the new population may be distinctively different, both genotypically and phenotypically, from the parent population from which it is derived. In extreme cases, the founder effect is thought to lead to the speciation and subsequent evolution of new species. In the figure shown, the original population has nearly equal numbers of blue and red individuals. The three smaller founder populations show that one or the other color may predominate (founder effect), due to random sampling of the original population. A population bottleneck may also cause a founder effect, though it is not strictly a new population. The founder effect o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Kilobase
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA and RNA. Dictated by specific hydrogen bonding patterns, "Watson–Crick" (or "Watson–Crick–Franklin") base pairs (guanine–cytosine and adenine– thymine) allow the DNA helix to maintain a regular helical structure that is subtly dependent on its nucleotide sequence. The complementary nature of this based-paired structure provides a redundant copy of the genetic information encoded within each strand of DNA. The regular structure and data redundancy provided by the DNA double helix make DNA well suited to the storage of genetic information, while base-pairing between DNA and incoming nucleotides provides the mechanism through which DNA polymerase replicates DNA and RNA polymerase transcribes DNA into RNA. Many DNA-binding protein ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
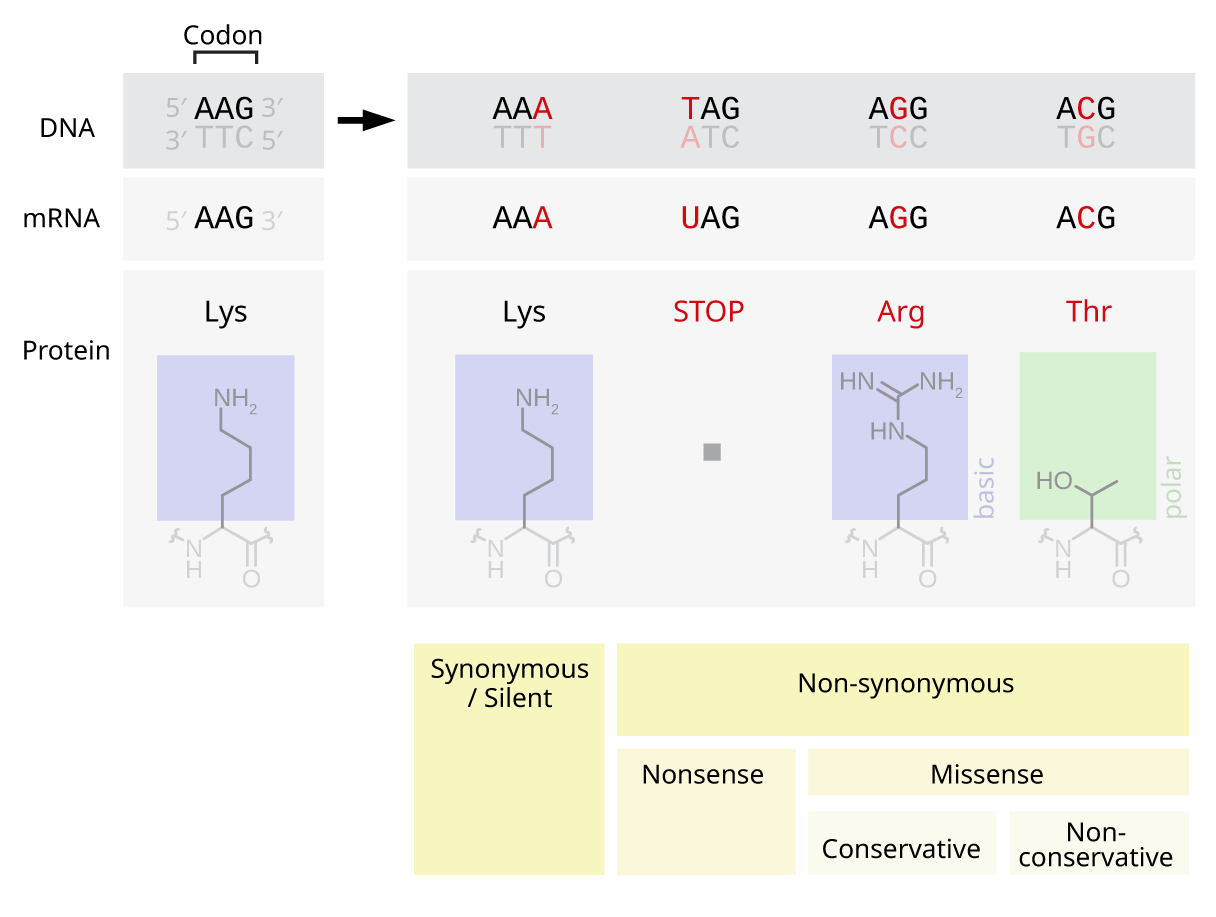
Point Mutation
A point mutation is a genetic mutation where a single nucleotide base is changed, inserted or deleted from a DNA or RNA sequence of an organism's genome. Point mutations have a variety of effects on the downstream protein product—consequences that are moderately predictable based upon the specifics of the mutation. These consequences can range from no effect (e.g. Synonymous substitution, synonymous mutations) to deleterious effects (e.g. frameshift mutations), with regard to protein production, composition, and function. Causes Point mutations usually take place during DNA replication. DNA replication occurs when one double-stranded DNA molecule creates two single strands of DNA, each of which is a template for the creation of the complementary strand. A single point mutation can change the whole DNA sequence. Changing one purine or pyrimidine may change the amino acid that the nucleotides code for. Point mutations may arise from spontaneous mutations that occur during DNA re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Germline
In biology and genetics, the germline is the population of a multicellular organism's cells that develop into germ cells. In other words, they are the cells that form gametes ( eggs and sperm), which can come together to form a zygote. They differentiate in the gonads from primordial germ cells into gametogonia, which develop into gametocytes, which develop into the final gametes. This process is known as gametogenesis. Germ cells pass on genetic material through the process of sexual reproduction. This includes fertilization, recombination and meiosis. These processes help to increase genetic diversity in offspring. Certain organisms reproduce asexually via processes such as apomixis, parthenogenesis, autogamy, and cloning. Apomixis and Parthenogenesis both refer to the development of an embryo without fertilization. The former typically occurs in plants seeds, while the latter tends to be seen in nematodes, as well as certain species of reptiles, birds, and fish. Autogamy ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |