|
Penicillin Binding Proteins
Penicillin-binding proteins (PBPs) are a group of proteins that are characterized by their affinity for and binding of penicillin. They are a normal constituent of many bacteria; the name just reflects the way by which the protein was discovered. All β-lactam antibiotics (except for tabtoxinine-β-lactam, which inhibits glutamine synthetase) bind to PBPs, which are essential for bacterial cell wall synthesis. PBPs are members of a subgroup of enzymes called transpeptidases. Specifically, PBPs are DD-transpeptidases. Diversity There are a large number of PBPs, usually several in each organism, and they are found as both membrane-bound and cytoplasmic proteins. For example, Spratt (1977) reports that six different PBPs are routinely detected in all strains of ''E. coli'' ranging in molecular weight from 40,000 to 91,000. The different PBPs occur in different numbers per cell and have varied affinities for penicillin. The PBPs are usually broadly classified into high-molecular ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Lysis
Lysis ( ) is the breaking down of the membrane of a cell, often by viral, enzymic, or osmotic (that is, "lytic" ) mechanisms that compromise its integrity. A fluid containing the contents of lysed cells is called a ''lysate''. In molecular biology, biochemistry, and cell biology laboratories, cell cultures may be subjected to lysis in the process of purifying their components, as in protein purification, DNA extraction, RNA extraction, or in purifying organelles. Many species of bacteria are subject to lysis by the enzyme lysozyme, found in animal saliva, egg white, and other secretions. Phage lytic enzymes (lysins) produced during bacteriophage infection are responsible for the ability of these viruses to lyse bacterial cells. Penicillin and related β-lactam antibiotics cause the death of bacteria through enzyme-mediated lysis that occurs after the drug causes the bacterium to form a defective cell wall. If the cell wall is completely lost and the penicillin was used ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Filamentation
Filamentation is the anomalous growth of certain bacteria, such as ''Escherichia coli'', in which cells continue to elongate but do not divide (no septa formation). The cells that result from elongation without division have multiple chromosomal copies. In the absence of antibiotics or other stressors, filamentation occurs at a low frequency in bacterial populations (4–8% short filaments and 0–5% long filaments in 1- to 8-hour cultures). The increased cell length can protect bacteria from protozoan predation and neutrophil phagocytosis by making ingestion of cells more difficult. Filamentation is also thought to protect bacteria from antibiotics, and is associated with other aspects of bacterial virulence such as biofilm formation. The number and length of filaments within a bacterial population increases when the bacteria are exposed to different physical, chemical and biological agents (e.g. UV light, DNA synthesis-inhibiting antibiotics, bacteriophages). This is termed ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
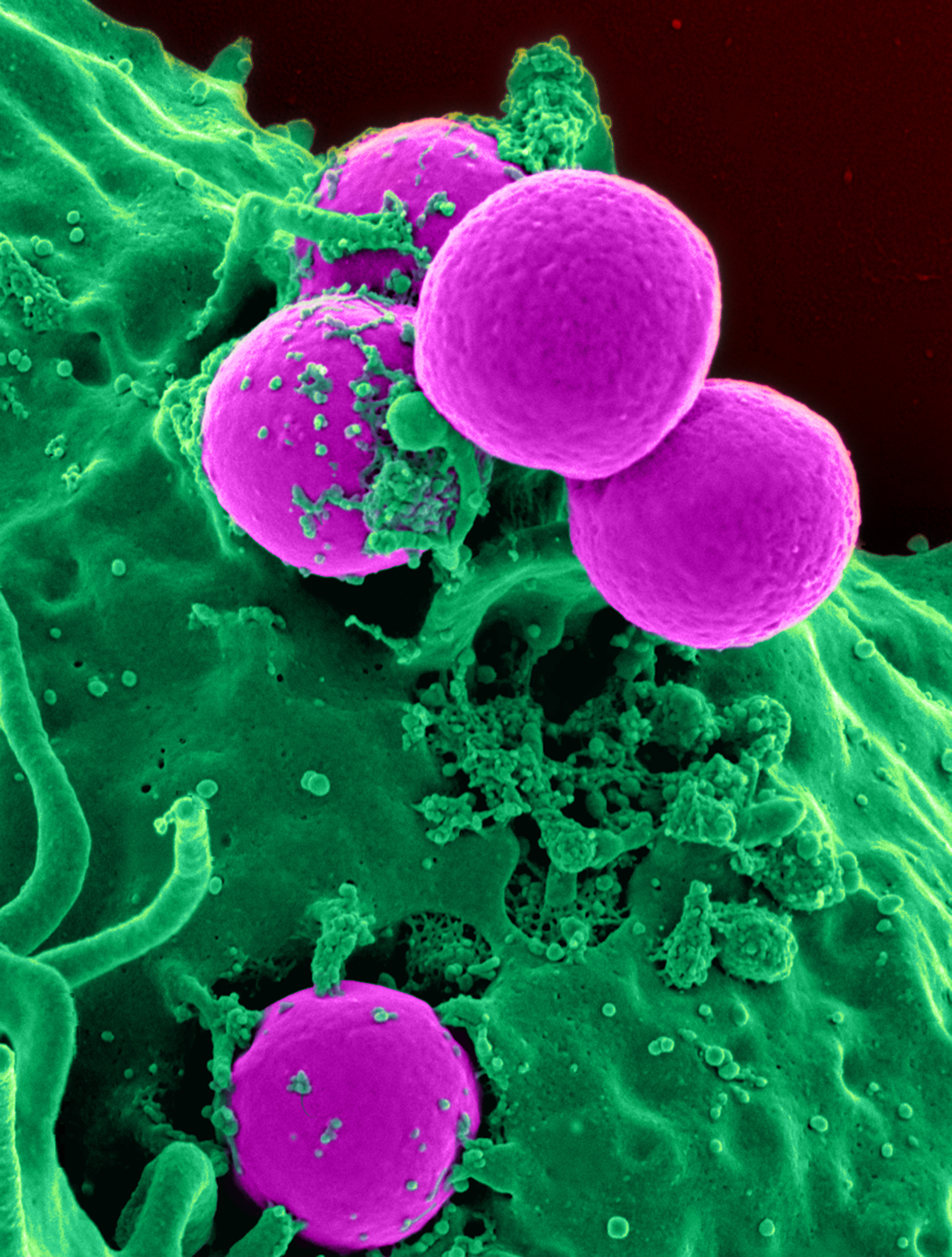
Methicillin-resistant Staphylococcus Aureus
Methicillin-resistant ''Staphylococcus aureus'' (MRSA) is a group of Gram-positive bacteria that are genetically distinct from other strains of ''Staphylococcus aureus''. MRSA is responsible for several difficult-to-treat infections in humans. It caused more than 100,000 deaths attributable to antimicrobial resistance in 2019. MRSA is any strain of ''S. aureus'' that has developed (through natural selection) or acquired (through horizontal gene transfer) a multiple drug resistance to beta-lactam antibiotics. Beta-lactam (β-lactam) antibiotics are a broad-spectrum group that include some penams (penicillin derivatives such as methicillin and oxacillin) and cephems such as the cephalosporins. Strains unable to resist these antibiotics are classified as methicillin-susceptible ''S. aureus'', or MSSA. MRSA is common in hospitals, prisons, and nursing homes, where people with open wounds, invasive devices such as catheters, and weakened immune systems are at greater risk of healt ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Antimicrobial Resistance
Antimicrobial resistance (AMR) occurs when microbes evolve mechanisms that protect them from the effects of antimicrobials. All classes of microbes can evolve resistance. Fungi evolve antifungal resistance. Viruses evolve antiviral resistance. Protozoa evolve antiprotozoal resistance, and bacteria evolve antibiotic resistance. Those bacteria that are considered extensively drug resistant (XDR) or totally drug-resistant (TDR) are sometimes called "superbugs".A.-P. Magiorakos, A. Srinivasan, R. B. Carey, Y. Carmeli, M. E. Falagas, C. G. Giske, S. Harbarth, J. F. Hinndler ''et al''Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria... Clinical Microbiology and Infection, Vol 8, Iss. 3 first published 27 July 2011 ia Wiley Online Library Retrieved 28 August 2020 Although antimicrobial resistance is a naturally-occurring process, it is often the result of improper usage of the drugs and management of the infections. Antibiotic resistance is a major subset o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Penicillin Binding Protein 2A
''mecA'' is a gene found in bacterial cells which allows them to be resistant to antibiotics such as methicillin, penicillin and other penicillin-like antibiotics. The bacteria strain most commonly known to carry ''mecA'' is methicillin-resistant ''Staphylococcus aureus'' (MRSA). In '' Staphylococcus'' species, ''mecA'' is spread through the staphylococcal chromosome cassette SCC''mec'' genetic element. Resistant strains cause many hospital-acquired infections. ''mecA'' encodes the protein PBP2A (penicillin-binding protein 2A), a transpeptidase that helps form the bacterial cell wall. PBP2A has a lower affinity for beta-lactam antibiotics such as methicillin and penicillin than DD-transpeptidase does, so it does not bind to the ringlike structure of penicillin-like antibiotics. This enables transpeptidase activity in the presence of beta-lactams, preventing them from inhibiting cell wall synthesis. The bacteria can then replicate as normal. History Methicillin resistance f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Lactamase
Beta-lactamases, (β-lactamases) are enzymes () produced by bacteria that provide multi-resistance to beta-lactam antibiotics such as penicillins, cephalosporins, cephamycins, monobactams and carbapenems (ertapenem), although carbapenems are relatively resistant to beta-lactamase. Beta-lactamase provides antibiotic resistance by breaking the antibiotics' structure. These antibiotics all have a common element in their molecular structure: a four-atom ring known as a beta-lactam (β-lactam) ring. Through hydrolysis, the enzyme lactamase breaks the β-lactam ring open, deactivating the molecule's antibacterial properties. Beta-lactam antibiotics are typically used to target a broad spectrum of gram-positive and gram-negative bacteria. Beta-lactamases produced by gram-negative bacteria are usually secreted, especially when antibiotics are present in the environment. Structure The structure of a '' Streptomyces'' serine β-lactamase (SBLs) is given by . The alpha-beta f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
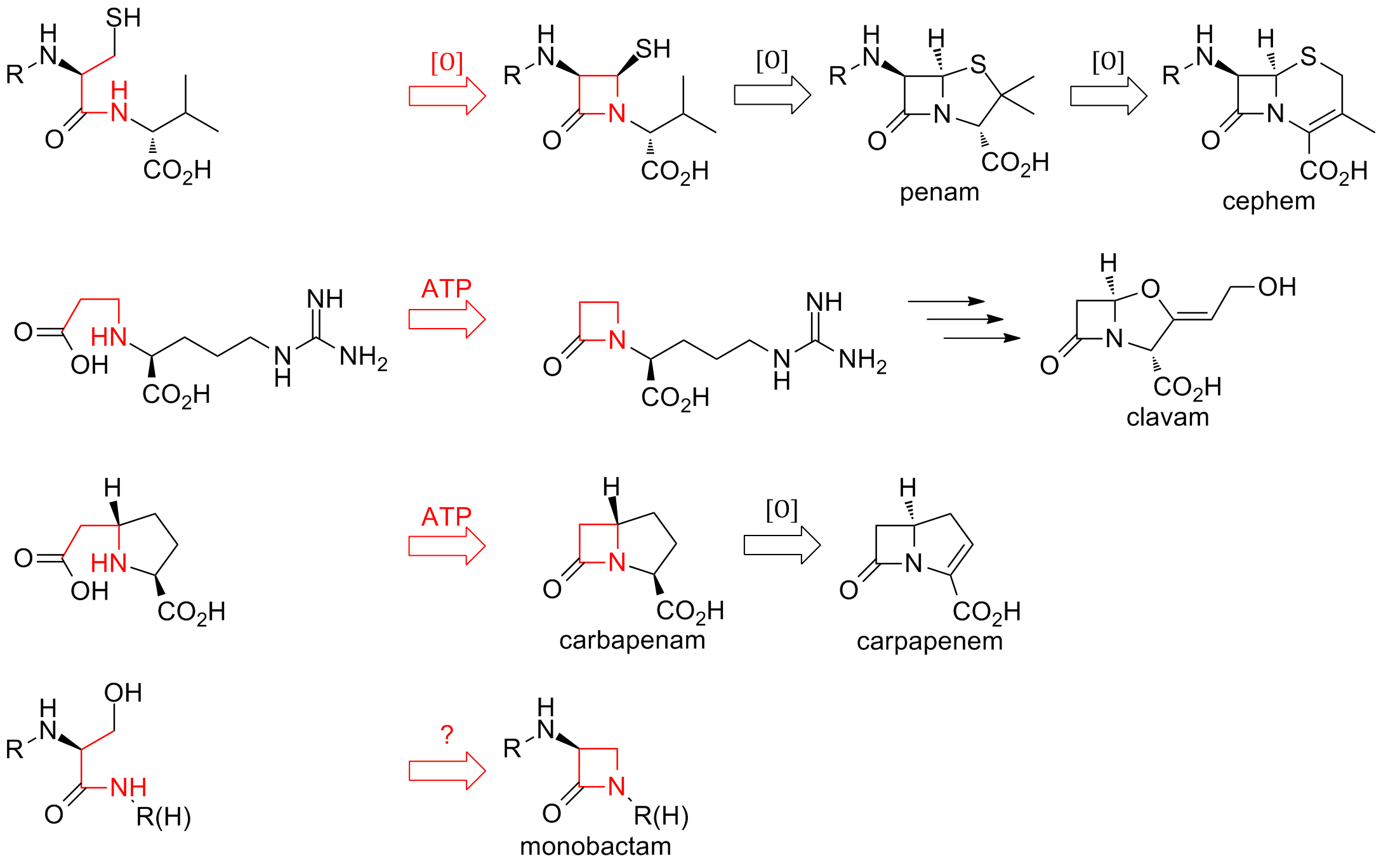
Β-lactam Antibiotic
β-lactam antibiotics (beta-lactam antibiotics) are antibiotics that contain a beta-lactam ring in their chemical structure. This includes penicillin derivatives (penams), cephalosporins and cephamycins (cephems), monobactams, carbapenems and carbacephems. Most β-lactam antibiotics work by inhibiting cell wall biosynthesis in the bacterial organism and are the most widely used group of antibiotics. Until 2003, when measured by sales, more than half of all commercially available antibiotics in use were β-lactam compounds. The first β-lactam antibiotic discovered, penicillin, was isolated from a strain of ''Penicillium rubens'' (named as ''Penicillium notatum'' at the time). Bacteria often develop resistance to β-lactam antibiotics by synthesizing a β-lactamase, an enzyme that attacks the β-lactam ring. To overcome this resistance, β-lactam antibiotics can be given with β-lactamase inhibitors such as clavulanic acid. Medical use β-lactam antibiotics are indicated fo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MreB
MreB is a protein found in bacteria that has been identified as a homologue of actin, as indicated by similarities in tertiary structure and conservation of active site peptide sequence. The conservation of protein structure suggests the common ancestry of the cytoskeletal elements formed by actin, found in eukaryotes, and MreB, found in prokaryotes. Indeed, recent studies have found that MreB proteins polymerize to form filaments that are similar to actin microfilaments. It has been shown to form multilayer sheets comprising diagonally interwoven filaments in the presence of ATP or GTP. MreB along with MreC and MreD are named after the mre operon (murein formation gene cluster E) to which they all belong. Function MreB controls the width of rod-shaped bacteria, such as ''Escherichia coli''. A mutant ''E. coli'' that creates defective MreB proteins will be spherical instead of rod-like. Also, most bacteria that are naturally spherical do not have the gene encoding MreB. Members ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
C-terminal End
The C-terminus (also known as the carboxyl-terminus, carboxy-terminus, C-terminal tail, C-terminal end, or COOH-terminus) is the end of an amino acid chain (protein or polypeptide), terminated by a free carboxyl group (-COOH). When the protein is translated from messenger RNA, it is created from N-terminus to C-terminus. The convention for writing peptide sequences is to put the C-terminal end on the right and write the sequence from N- to C-terminus. Chemistry Each amino acid has a carboxyl group and an amine group. Amino acids link to one another to form a chain by a dehydration reaction which joins the amine group of one amino acid to the carboxyl group of the next. Thus polypeptide chains have an end with an unbound carboxyl group, the C-terminus, and an end with an unbound amine group, the N-terminus. Proteins are naturally synthesized starting from the N-terminus and ending at the C-terminus. Function C-terminal retention signals While the N-terminus of a protein often conta ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
N-terminal End
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the amine group is bonded to the carboxylic group of another amino acid, making it a chain. That leaves a free carboxylic group at one end of the peptide, called the C-terminus, and a free amine group on the other end called the N-terminus. By convention, peptide sequences are written N-terminus to C-terminus, left to right (in LTR writing systems). This correlates the translation direction to the text direction, because when a protein is translated from messenger RNA, it is created from the N-terminus to the C-terminus, as amino acids are added to the carboxyl end of the protein. Chemistry Each amino acid has an amine group and a carboxylic group. Amino acids link to one another by peptide bonds which form through a dehydration reaction that jo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are catalytic RNA molecules, called ribozymes. Enzymes' specificity comes from their unique three-dimensional structures. Like all catalysts, enzymes increase the reaction ra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |