|
NrDNA
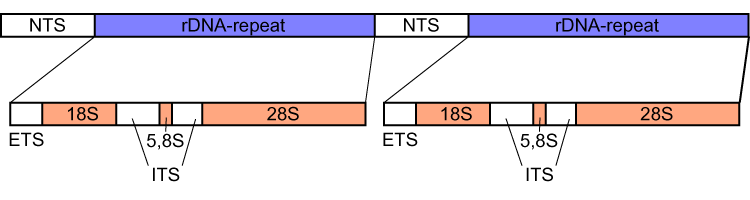
Ribosomal DNA (rDNA) is a DNA sequence that codes for ribosomal RNA. These sequences regulate transcription initiation and amplification, and contain both transcribed and non-transcribed spacer segments. In the human genome there are 5 chromosomes with nucleolus organizer regions: the acrocentric chromosomes 13 (RNR1), 14 ( RNR2), 15 ( RNR3), 21 (RNR4) and 22 (RNR5). The genes that are responsible for encoding the various sub-units of rRNA are located across multiple chromosomes in humans. But the genes that encode for rRNA are highly conserved across the domains, with only the copy numbers involved for the genes having varying numbers per species. In Bacteria, Archaea, and chloroplasts the rRNA is composed of different (smaller) units, the large (23S) ribosomal RNA, 16S ribosomal RNA and 5S rRNA. The 16S rRNA is widely used for phylogenetic studies. Eukaryotes The rRNA transcribed from the approximately 600 rDNA repeats forms the most abundant section of RNA found in cell ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phylogenetics
In biology, phylogenetics (; from Greek φυλή/ φῦλον [] "tribe, clan, race", and wikt:γενετικός, γενετικός [] "origin, source, birth") is the study of the evolutionary history and relationships among or within groups of organisms. These relationships are determined by Computational phylogenetics, phylogenetic inference methods that focus on observed heritable traits, such as DNA sequences, protein amino acid sequences, or morphology. The result of such an analysis is a phylogenetic tree—a diagram containing a hypothesis of relationships that reflects the evolutionary history of a group of organisms. The tips of a phylogenetic tree can be living taxa or fossils, and represent the "end" or the present time in an evolutionary lineage. A phylogenetic diagram can be rooted or unrooted. A rooted tree diagram indicates the hypothetical common ancestor of the tree. An unrooted tree diagram (a network) makes no assumption about the ancestral line, and do ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleolus Organizer Region
] Nucleolus organizer regions (NORs) are chromosome, chromosomal regions crucial for the formation of the nucleolus. In humans, the NORs are located on the short arms of the acrocentric chromosomes 13, 14, 15, 21 and 22, the genes RNR1, RNR2, RNR3, RNR4, and RNR5 respectively. These regions code for 5.8S, 18S, and 28S ribosomal RNA. The NORs are "sandwiched" between the repetitive, heterochromatic DNA sequences of the centromeres and telomeres. The exact sequence of these regions is not included in the human reference genome as of 2016 or the GRCh38.p10 released January 6, 2017. On 28 February 2019, GRCh38.p13 was released, which added the NOR sequences for the short arms of chromosomes 13, 14, 15, 21, and 22. However, it is known that NORs contain tandem copies of ribosomal DNA (rDNA) genes. Some sequences of flanking sequences proximal and distal to NORs have been reported. The NORs of a loris have been reported to be highly variable. There are also DNA sequences relate ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleolus
The nucleolus (, plural: nucleoli ) is the largest structure in the nucleus of eukaryotic cells. It is best known as the site of ribosome biogenesis, which is the synthesis of ribosomes. The nucleolus also participates in the formation of signal recognition particles and plays a role in the cell's response to stress. Nucleoli are made of proteins, DNA and RNA, and form around specific chromosomal regions called nucleolar organizing regions. Malfunction of nucleoli can be the cause of several human conditions called "nucleolopathies" and the nucleolus is being investigated as a target for cancer chemotherapy. History The nucleolus was identified by bright-field microscopy during the 1830s. Little was known about the function of the nucleolus until 1964, when a study of nucleoli by John Gurdon and Donald Brown in the African clawed frog ''Xenopus laevis'' generated increasing interest in the function and detailed structure of the nucleolus. They found that 25% of the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Drosophila
''Drosophila'' () is a genus of flies, belonging to the family Drosophilidae, whose members are often called "small fruit flies" or (less frequently) pomace flies, vinegar flies, or wine flies, a reference to the characteristic of many species to linger around overripe or rotting fruit. They should not be confused with the Tephritidae, a related family, which are also called fruit flies (sometimes referred to as "true fruit flies"); tephritids feed primarily on unripe or ripe fruit, with many species being regarded as destructive agricultural pests, especially the Mediterranean fruit fly. One species of ''Drosophila'' in particular, '' D. melanogaster'', has been heavily used in research in genetics and is a common model organism in developmental biology. The terms "fruit fly" and "''Drosophila''" are often used synonymously with ''D. melanogaster'' in modern biological literature. The entire genus, however, contains more than 1,500 species and is very diverse in appearan ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
5S Ribosomal RNA
The 5S ribosomal RNA (5S rRNA) is an approximately 120 nucleotide-long ribosomal RNA molecule with a mass of 40 kDa. It is a structural and functional component of the large subunit of the ribosome in all domains of life (bacteria, archaea, and eukaryotes), with the exception of mitochondrial ribosomes of fungi and animals. The designation 5S refers to the molecule's sedimentation velocity in an ultracentrifuge, which is measured in Svedberg units (S). Biosynthesis In prokaryotes, the 5S rRNA gene is typically located in the rRNA operons downstream of the small and the large subunit rRNA, and co-transcribed into a polycistronic precursor. A particularity of eukaryotic nuclear genomes is the occurrence of multiple 5S rRNA gene copies (5S rDNA) clustered in tandem repeats, with copy number varying from species to species. Eukaryotic 5S rRNA is synthesized by RNA polymerase III, whereas other eukaryotic rRNAs are cleaved from a 45S precursor transcribed by RNA polymerase I ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ITS2
Internal transcribed spacer (ITS) is the spacer DNA situated between the small-subunit ribosomal RNA (rRNA) and large-subunit rRNA genes in the chromosome or the corresponding transcribed region in the polycistronic rRNA precursor transcript. ITS across life domains In bacteria and archaea, there is a single ITS, located between the 16S and 23S rRNA genes. Conversely, there are two ITSs in eukaryotes: ITS1 is located between 18S and 5.8S rRNA genes, while ITS2 is between 5.8S and 28S (in opisthokonts, or 25S in plants) rRNA genes. ITS1 corresponds to the ITS in bacteria and archaea, while ITS2 originated as an insertion that interrupted the ancestral 23S rRNA gene. Organization In bacteria and archaea, the ITS occurs in one to several copies, as do the flanking 16S and 23S genes. When there are multiple copies, these do not occur adjacent to one another. Rather, they occur in discrete locations in the circular chromosome. It is not uncommon in bacteria to carry tRN ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ITS1
Internal transcribed spacer (ITS) is the spacer DNA situated between the small-subunit ribosomal RNA (rRNA) and large-subunit rRNA genes in the chromosome or the corresponding transcribed region in the polycistronic rRNA precursor transcript. ITS across life domains In bacteria and archaea, there is a single ITS, located between the 16S and 23S rRNA genes. Conversely, there are two ITSs in eukaryotes: ITS1 is located between 18S and 5.8S rRNA genes, while ITS2 is between 5.8S and 28S (in opisthokonts, or 25S in plants) rRNA genes. ITS1 corresponds to the ITS in bacteria and archaea, while ITS2 originated as an insertion that interrupted the ancestral 23S rRNA gene. Organization In bacteria and archaea, the ITS occurs in one to several copies, as do the flanking 16S and 23S genes. When there are multiple copies, these do not occur adjacent to one another. Rather, they occur in discrete locations in the circular chromosome. It is not uncommon in bacteria to carry tRN ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
18S Ribosomal RNA
18S ribosomal RNA (abbreviated 18S rRNA) is a part of the ribosomal RNA. The S in 18S represents Svedberg units. 18S rRNA is an SSU rRNA, a component of the eukaryotic ribosomal small subunit ( 40S). 18S rRNA is the structural RNA for the small component of eukaryotic cytoplasmic ribosomes, and thus one of the basic components of all eukaryotic cells. 18S rRNA is the eukaryotic cytosolic homologue of 16S ribosomal RNA in prokaryotes and plastids. 18S rRNA is also a homologue of 12S ribosomal RNA in mitochondria. The genes coding for 18S rRNA are referred to as 18S rRNA genes. Sequence data from these genes is widely used in molecular analysis to reconstruct the evolutionary history of organisms, especially in vertebrates, as its slow evolutionary rate makes it suitable to reconstruct ancient divergences. Uses in phylogeny The small subunit (SSU) 18S rRNA gene is one of the most frequently used genes in phylogenetic studies and an important marker for random target polymera ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tandemly Arrayed Genes
Tandemly arrayed genes (TAGs) are a gene cluster created by tandem duplications, a process in which one gene is duplicated and the copy is found adjacent to the original. They serve to encode large numbers of genes at a time. TAGs represent a large proportion of genes in a genome, including between 14% to 17% of the human, mouse, and rat genomes. TAG clusters may have as few as two genes, with small clusters predominating, but may consist of hundreds of genes. An example are tandem clusters of rRNA encoding genes. These genes are transcribed faster than they would be if only a single copy of the gene was available. Additionally, a single RNA gene may not be able to provide enough RNA, but tandem repeats of the gene allow sufficient RNA to be produced. For example, cells in a human embryo contain between five and ten million ribosomes, and cell number doubles within 24 hours. In order to provide the necessary ribosomes, multiple RNA polymerases must consecutively transcribe mul ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Messenger RNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein. mRNA is created during the process of transcription, where an enzyme ( RNA polymerase) converts the gene into primary transcript mRNA (also known as pre-mRNA). This pre-mRNA usually still contains introns, regions that will not go on to code for the final amino acid sequence. These are removed in the process of RNA splicing, leaving only exons, regions that will encode the protein. This exon sequence constitutes mature mRNA. Mature mRNA is then read by the ribosome, and, utilising amino acids carried by transfer RNA (tRNA), the ribosome creates the protein. This process is known as translation. All of these processes form part of the central dogma of molecular biology, which describes the flow of genetic information in a biological system. As in DNA, gen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Translation (biology)
In molecular biology and genetics, translation is the process in which ribosomes in the cytoplasm or endoplasmic reticulum synthesize proteins after the process of transcription of DNA to RNA in the cell's nucleus. The entire process is called gene expression. In translation, messenger RNA (mRNA) is decoded in a ribosome, outside the nucleus, to produce a specific amino acid chain, or polypeptide. The polypeptide later folds into an active protein and performs its functions in the cell. The ribosome facilitates decoding by inducing the binding of complementary tRNA anticodon sequences to mRNA codons. The tRNAs carry specific amino acids that are chained together into a polypeptide as the mRNA passes through and is "read" by the ribosome. Translation proceeds in three phases: # Initiation: The ribosome assembles around the target mRNA. The first tRNA is attached at the start codon. # Elongation: The last tRNA validated by the small ribosomal subunit (''acc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
.png)