|
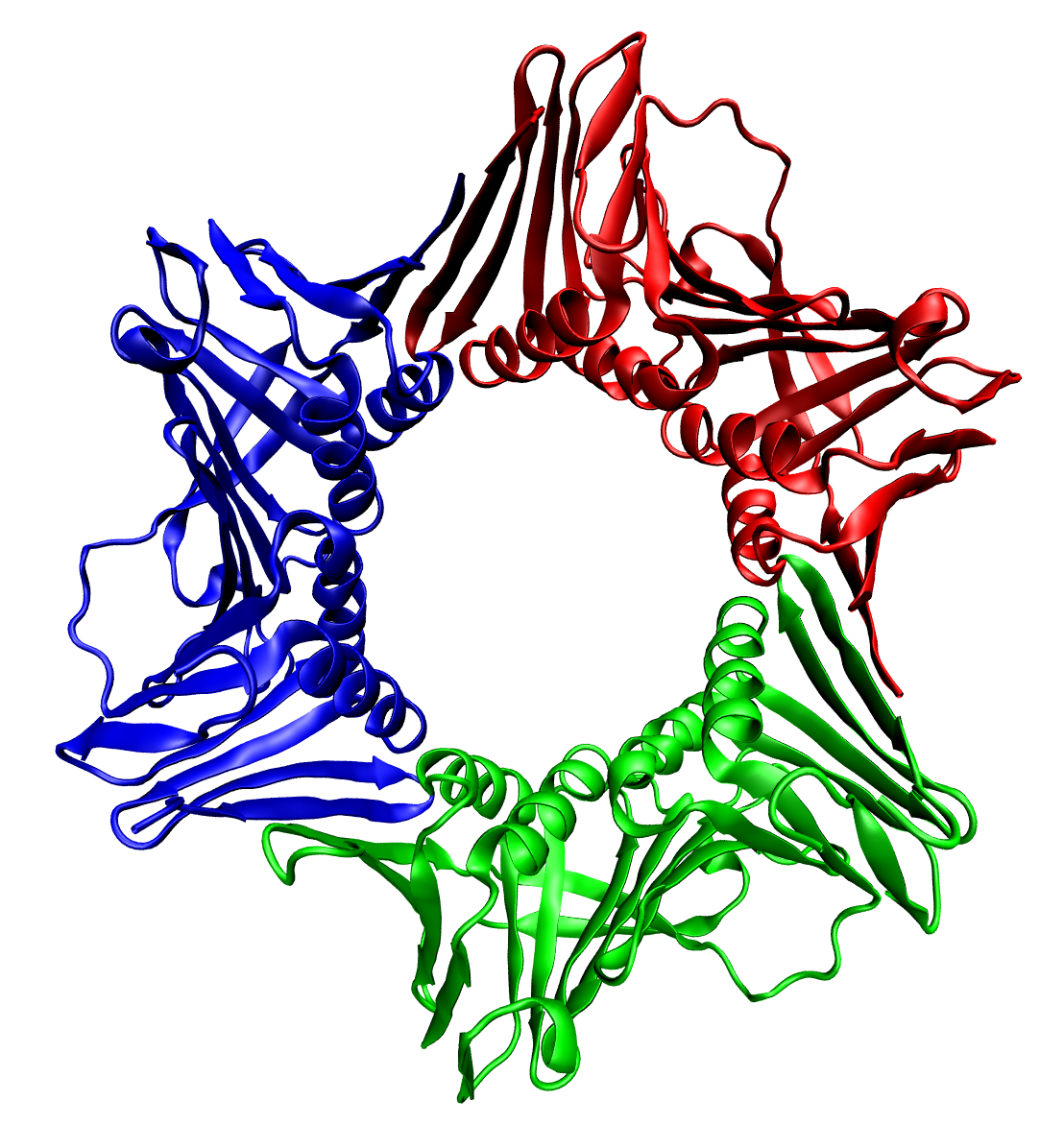
Multienzyme Complex
Multienzyme complex contains several copies of one or several enzymes (polypeptide chains) packed into one assembly. Multienzyme complex carries out a single or a series of biochemical reactions taking place in the cells. It allows to segregate certain biochemical pathways into one place in the cell. Examples include pyruvate dehydrogenase, fatty acid synthetase, glutamine synthetase, proteasome, rubisco. See also * Quaternary structure * Protein complex * Macromolecular assembly * Biomolecular complex A biomolecule or biological molecule is a loosely used term for molecules present in organisms that are essential to one or more typically biological processes, such as cell division, morphogenesis, or development. Biomolecules include large ... References {{reflist Enzymes ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are catalytic RNA molecules, called ribozymes. Enzymes' specificity comes from their unique three-dimensional structures. Like all catalysts, enzymes increase the reaction ra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polypeptide Chains
Peptides (, ) are short chains of amino acids linked by peptide bonds. Long chains of amino acids are called proteins. Chains of fewer than twenty amino acids are called oligopeptides, and include dipeptides, tripeptides, and tetrapeptides. A polypeptide is a longer, continuous, unbranched peptide chain. Hence, peptides fall under the broad chemical classes of biological polymers and oligomers, alongside nucleic acids, oligosaccharides, polysaccharides, and others. A polypeptide that contains more than approximately 50 amino acids is known as a protein. Proteins consist of one or more polypeptides arranged in a biologically functional way, often bound to ligands such as coenzymes and cofactors, or to another protein or other macromolecule such as DNA or RNA, or to complex macromolecular assemblies. Amino acids that have been incorporated into peptides are termed residues. A water molecule is released during formation of each amide bond.. All peptides except cyclic peptides ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
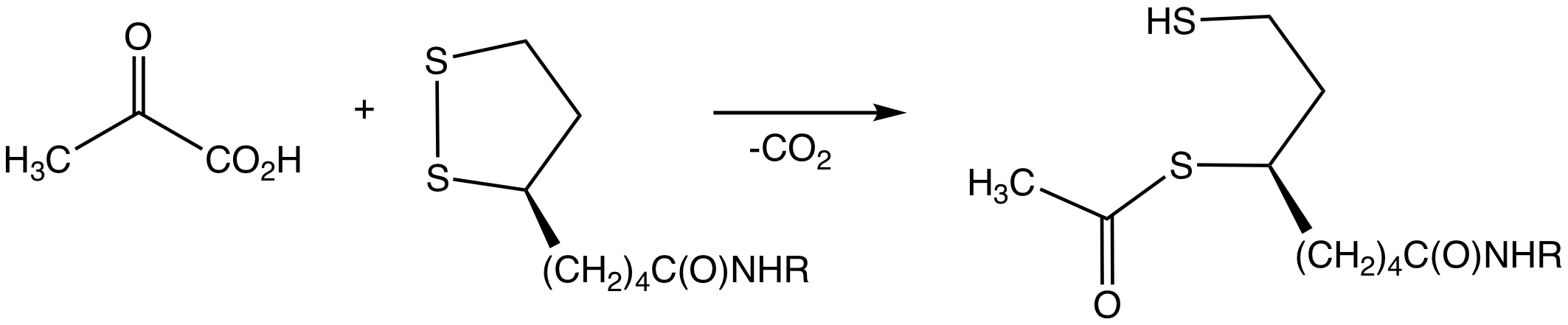
Pyruvate Dehydrogenase
Pyruvate dehydrogenase is an enzyme that catalyzes the reaction of pyruvate and a lipoamide to give the acetylated dihydrolipoamide and carbon dioxide. The conversion requires the coenzyme thiamine pyrophosphate. Pyruvate dehydrogenase is usually encountered as a component, referred to as E1, of the pyruvate dehydrogenase complex (PDC). PDC consists of other enzymes, referred to as E2 and E3. Collectively E1-E3 transform pyruvate, NAD+, coenzyme A into acetyl-CoA, CO2, and NADH. The conversion is crucial because acetyl-CoA may then be used in the citric acid cycle to carry out cellular respiration. To distinguish between this enzyme and the PDC, it is systematically called pyruvate dehydrogenase (acetyl-transferring). Mechanism The thiamine pyrophosphate (TPP) converts to an ylide by deprotonation. The ylide attack the ketone group of pyruvate. The resulting adduct decarboxylates. The resulting 1,3-dipole reductively acetylates lipoamide-E2. In terms of details, bioch ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fatty Acid Synthetase
Fatty acid synthase (FAS) is an enzyme that in humans is encoded by the ''FASN'' gene. Fatty acid synthase is a multi-enzyme protein that catalyzes fatty acid synthesis. It is not a single enzyme but a whole enzymatic system composed of two identical 272 kDa multifunctional polypeptides, in which substrates are handed from one functional domain to the next. Its main function is to catalyze the synthesis of palmitate (C16:0, a long-chain saturated fatty acid) from acetyl-CoA and malonyl-CoA, in the presence of NADPH. The fatty acids are synthesized by a series of decarboxylative Claisen condensation reactions from acetyl-CoA and malonyl-CoA. Following each round of elongation the beta keto group is reduced to the fully saturated carbon chain by the sequential action of a ketoreductase (KR), dehydratase (DH), and enoyl reductase (ER). The growing fatty acid chain is carried between these active sites while attached covalently to the phosphopantetheine prosthetic group of an acy ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Glutamine Synthetase
Glutamine synthetase (GS) () is an enzyme that plays an essential role in the metabolism of nitrogen by catalyzing the condensation of glutamate and ammonia to form glutamine: Glutamate + Adenosine triphosphate, ATP + NH3 → Glutamine + Adenosine diphosphate, ADP + phosphate Glutamine synthetase uses ammonia produced by nitrate reduction, amino acid degradation, and photorespiration. The amide group of glutamate is a nitrogen source for the synthesis of glutamine pathway metabolites. Other reactions may take place via GS. Competition between ammonium ion and water, their binding affinities, and the concentration of ammonium ion, influences glutamine synthesis and glutamine hydrolysis. Glutamine is formed if an ammonium ion attacks the acyl-phosphate intermediate, while glutamate is remade if water attacks the intermediate. Ammonium ion binds more strongly than water to GS due to electrostatic forces between a cation and a negatively charged pocket. Another possible reac ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Proteasome
Proteasomes are protein complexes which degrade unneeded or damaged proteins by proteolysis, a chemical reaction that breaks peptide bonds. Enzymes that help such reactions are called proteases. Proteasomes are part of a major mechanism by which cells regulate the concentration of particular proteins and degrade misfolded proteins. Proteins are tagged for degradation with a small protein called ubiquitin. The tagging reaction is catalyzed by enzymes called ubiquitin ligases. Once a protein is tagged with a single ubiquitin molecule, this is a signal to other ligases to attach additional ubiquitin molecules. The result is a ''polyubiquitin chain'' that is bound by the proteasome, allowing it to degrade the tagged protein. The degradation process yields peptides of about seven to eight amino acids long, which can then be further degraded into shorter amino acid sequences and used in synthesizing new proteins. Proteasomes are found inside all eukaryotes and archaea, and in so ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Rubisco
Ribulose-1,5-bisphosphate carboxylase-oxygenase, commonly known by the abbreviations RuBisCo, rubisco, RuBPCase, or RuBPco, is an enzyme () involved in the first major step of carbon fixation, a process by which atmospheric carbon dioxide is converted by plants and other photosynthesis, photosynthetic organisms to fuel, energy-rich molecules such as glucose. In chemical terms, it catalysis, catalyzes the carboxylation of ribulose-1,5-bisphosphate (also known as RuBP). It is probably the most abundant enzyme on Earth. Alternative carbon fixation pathways RuBisCO is important biology, biologically because it catalyzes the primary chemical reaction by which Total inorganic carbon, inorganic carbon enters the biosphere. While many autotrophic bacteria and archaea fix carbon via the reductive acetyl CoA Pathway, reductive acetyl CoA pathway, the 3-hydroxypropionate cycle, or the reverse Krebs cycle, these pathways are relatively small contributors to global carbon fixation compared t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Quaternary Structure
Protein quaternary structure is the fourth (and highest) classification level of protein structure. Protein quaternary structure refers to the structure of proteins which are themselves composed of two or more smaller protein chains (also referred to as subunits). Protein quaternary structure describes the number and arrangement of multiple folded protein subunits in a multi-subunit complex. It includes organizations from simple dimers to large homooligomers and complexes with defined or variable numbers of subunits. In contrast to the first three levels of protein structure, not all proteins will have a quaternary structure since some proteins function as single units. Protein quaternary structure can also refer to biomolecular complexes of proteins with nucleic acids and other cofactors. Description and examples Many proteins are actually assemblies of multiple polypeptide chains. The quaternary structure refers to the number and arrangement of the protein subunit ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Complex
A protein complex or multiprotein complex is a group of two or more associated polypeptide chains. Protein complexes are distinct from multienzyme complexes, in which multiple catalytic domains are found in a single polypeptide chain. Protein complexes are a form of quaternary structure. Proteins in a protein complex are linked by non-covalent protein–protein interactions. These complexes are a cornerstone of many (if not most) biological processes. The cell is seen to be composed of modular supramolecular complexes, each of which performs an independent, discrete biological function. Through proximity, the speed and selectivity of binding interactions between enzymatic complex and substrates can be vastly improved, leading to higher cellular efficiency. Many of the techniques used to enter cells and isolate proteins are inherently disruptive to such large complexes, complicating the task of determining the components of a complex. Examples of protein complexes include the p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Macromolecular Assembly
The term macromolecular assembly (MA) refers to massive chemical structures such as viruses and non-biologic nanoparticles, cellular organelles and membranes and ribosomes, etc. that are complex mixtures of polypeptide, polynucleotide, polysaccharide or other polymeric macromolecules. They are generally of more than one of these types, and the mixtures are defined spatially (i.e., with regard to their chemical shape), and with regard to their underlying chemical composition and structure. Macromolecules are found in living and nonliving things, and are composed of many hundreds or thousands of atoms held together by covalent bonds; they are often characterized by repeating units (i.e., they are polymers). Assemblies of these can likewise be biologic or non-biologic, though the MA term is more commonly applied in biology, and the term supramolecular assembly is more often applied in non-biologic contexts (e.g., in supramolecular chemistry and nanotechnology). MAs of macromolecules ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Biomolecular Complex
A biomolecule or biological molecule is a loosely used term for molecules present in organisms that are essential to one or more typically biological processes, such as cell division, morphogenesis, or development. Biomolecules include large macromolecules (or polyelectrolytes) such as proteins, carbohydrates, lipids, and nucleic acids, as well as small molecules such as primary metabolites, secondary metabolites and natural products. A more general name for this class of material is biological materials. Biomolecules are an important element of living organisms, those biomolecules are often endogenous, produced within the organism but organisms usually need exogenous biomolecules, for example certain nutrients, to survive. Biology and its subfields of biochemistry and molecular biology study biomolecules and their reactions. Most biomolecules are organic compounds, and just four elements—oxygen, carbon, hydrogen, and nitrogen—make up 96% of the human body's mass. Bu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |