|
Metatranscriptomics
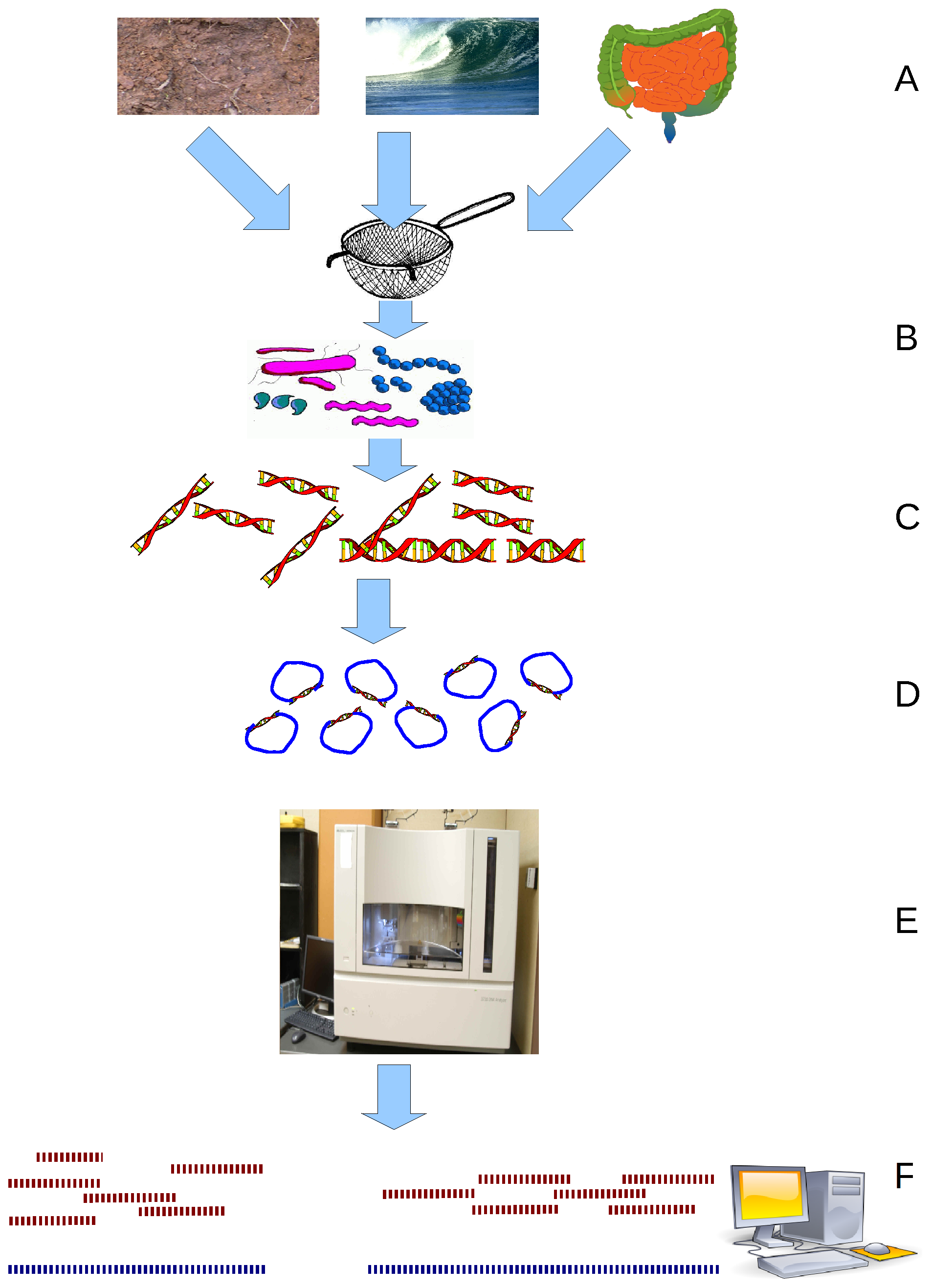
Metatranscriptomics is the science that studies gene expression of microbes within natural environments, i.e., the metatranscriptome. It also allows to obtain whole gene expression profiling of complex microbial communities. While metagenomics focuses on studying the genomic content and on identifying which microbes are present within a community, metatranscriptomics can be used to study the diversity of the active genes within such community, to quantify their expression levels and to monitor how these levels change in different conditions (e.g., physiological vs. pathological conditions in an organism). The advantage of metatranscriptomics is that it can provide information about differences in the active functions of microbial communities which appear to be the same in terms of microbe composition. Introduction The microbiome has been defined as a microbial community occupying a well-defined habitat. They are ubiquitous and extremely relevant for the maintenance of” the chara ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Metagenomics
Metagenomics is the study of genetic material recovered directly from environmental or clinical samples by a method called sequencing. The broad field may also be referred to as environmental genomics, ecogenomics, community genomics or microbiomics. While traditional microbiology and microbial genome sequencing and genomics rely upon cultivated clonal cultures, early environmental gene sequencing cloned specific genes (often the 16S rRNA gene) to produce a profile of diversity in a natural sample. Such work revealed that the vast majority of microbial biodiversity had been missed by cultivation-based methods. Because of its ability to reveal the previously hidden diversity of microscopic life, metagenomics offers a powerful lens for viewing the microbial world that has the potential to revolutionize understanding of the entire living world. As the price of DNA sequencing continues to fall, metagenomics now allows microbial ecology to be investigated at a much greater scale ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene
In biology, the word gene (from , ; "...Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a basic unit of heredity and the molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protein-coding genes and noncoding genes. During gene expression, the DNA is first copied into RNA. The RNA can be directly functional or be the intermediate template for a protein that performs a function. The transmission of genes to an organism's offspring is the basis of the inheritance of phenotypic traits. These genes make up different DNA sequences called genotypes. Genotypes along with environmental and developmental factors determine what the phenotypes will be. Most biological traits are under the influence of polygenes (many different genes) as well as gen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
BLAST (biotechnology)
In bioinformatics, BLAST (basic local alignment search tool) is an algorithm and program for comparing primary biological sequence information, such as the amino-acid sequences of proteins or the nucleotides of DNA and/or RNA sequences. A BLAST search enables a researcher to compare a subject protein or nucleotide sequence (called a query) with a library or database of sequences, and identify database sequences that resemble alphabet above a certain threshold. For example, following the discovery of a previously unknown gene in the mouse, a scientist will typically perform a BLAST search of the human genome to see if humans carry a similar gene; BLAST will identify sequences in the pig genome that resemble the mouse gene based on similarity of sequence. Background BLAST, which ''The New York Times'' called ''the Google of biological research'', is one of the most widely used bioinformatics programs for sequence searching. It addresses a fundamental problem in bioinformatics r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Illumina Dye Sequencing
Illumina dye sequencing is a technique used to determine the series of base pairs in DNA, also known as DNA sequencing. The reversible terminated chemistry concept was invented by Bruno Canard and Simon Sarfati at the Pasteur Institute in Paris. It was developed by Shankar Balasubramanian and David Klenerman of Cambridge University, who subsequently founded Solexa, a company later acquired by Illumina. This sequencing method is based on reversible dye-terminators that enable the identification of single nucleotides as they are washed over DNA strands. It can also be used for whole-genome and region sequencing, transcriptome analysis, metagenomics, small RNA discovery, methylation profiling, and genome-wide protein- nucleic acid interaction analysis. Overview This works in three basic steps: amplify, sequence, and analyze. The process begins with purified DNA. The DNA is fragmented and adapters are added that contain segments that act as reference points during amplification, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Amino Acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although hundreds of amino acids exist in nature, by far the most important are the alpha-amino acids, which comprise proteins. Only 22 alpha amino acids appear in the genetic code. Amino acids can be classified according to the locations of the core structural functional groups, as Alpha and beta carbon, alpha- , beta- , gamma- or delta- amino acids; other categories relate to Chemical polarity, polarity, ionization, and side chain group type (aliphatic, Open-chain compound, acyclic, aromatic, containing hydroxyl or sulfur, etc.). In the form of proteins, amino acid '' residues'' form the second-largest component (water being the largest) of human muscles and other tissues. Beyond their role as residues in proteins, amino acids participate in a number of processes such as neurotransmitter transport and biosynthesis. It is thought that they played a key role in enabling life ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MG-RAST
MG-RAST is an open-source web application server that suggests automatic phylogenetic and functional analysis of metagenomes. It is also one of the biggest repositories for metagenomic data. The name is an abbreviation of ''Metagenomic Rapid Annotations using Subsystems Technology''. The pipeline automatically produces functional assignments to the sequences that belong to the metagenome by performing sequence comparisons to databases in both nucleotide and amino-acid levels. The applications supply phylogenetic and functional assignments of the metagenome being analysed, as well as tools for comparing different metagenomes. It also provides RESTful APIfor programmatic access. The server was created and maintained by Argonne National Laboratory from the University of Chicago. In December 29 of 2016, the system had analyzed 60 terabase-pairs of data from more than 150,000 data sets. Among the analyzed data sets, more than 23,000 are available to the public. Currently, the computa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
16S Ribosomal RNA
16 S ribosomal RNA (or 16 S rRNA) is the RNA component of the 30S subunit of a prokaryotic ribosome (SSU rRNA). It binds to the Shine-Dalgarno sequence and provides most of the SSU structure. The genes coding for it are referred to as 16S rRNA gene and are used in reconstructing phylogenies, due to the slow rates of evolution of this region of the gene. Carl Woese and George E. Fox were two of the people who pioneered the use of 16S rRNA in phylogenetics in 1977. Multiple sequences of the 16S rRNA gene can exist within a single bacterium. Functions * Like the large (23S) ribosomal RNA, it has a structural role, acting as a scaffold defining the positions of the ribosomal proteins. * The 3-end contains the anti- Shine-Dalgarno sequence, which binds upstream to the AUG start codon on the mRNA. The 3-end of 16S RNA binds to the proteins S1 and S21 which are known to be involved in initiation of protein synthesis * Interacts with 23S, aiding in the binding of the two ribosomal s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Human Microbiome Project
The Human Microbiome Project (HMP) was a United States National Institutes of Health (NIH) research initiative to improve understanding of the microbiota involved in human health and disease. Launched in 2007, the first phase (HMP1) focused on identifying and characterizing human microbiota. The second phase, known as the Integrative Human Microbiome Project (iHMP) launched in 2014 with the aim of generating resources to characterize the microbiome and elucidating the roles of microbes in health and disease states. The program received $170 million in funding by the NIH Common Fund from 2007 to 2016. Important components of the HMP were culture-independent methods of microbial community characterization, such as metagenomics (which provides a broad genetic perspective on a single microbial community), as well as extensive whole genome sequencing (which provides a "deep" genetic perspective on certain aspects of a given microbial community, ''i.e.'' of individual bacterial specie ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA-seq
RNA-Seq (named as an abbreviation of RNA sequencing) is a sequencing technique which uses next-generation sequencing (NGS) to reveal the presence and quantity of RNA in a biological sample at a given moment, analyzing the continuously changing cellular transcriptome. Specifically, RNA-Seq facilitates the ability to look at alternative gene spliced transcripts, post-transcriptional modifications, gene fusion, mutations/SNPs and changes in gene expression over time, or differences in gene expression in different groups or treatments. In addition to mRNA transcripts, RNA-Seq can look at different populations of RNA to include total RNA, small RNA, such as miRNA, tRNA, and ribosomal profiling. RNA-Seq can also be used to determine exon/intron boundaries and verify or amend previously annotated 5' and 3' gene boundaries. Recent advances in RNA-Seq include single cell sequencing, in situ sequencing of fixed tissue, and native RNA molecule sequencing with single-molecule real-time ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
De Novo Transcriptome Assembly
''De novo'' transcriptome assembly is the de novo sequence assembly method of creating a transcriptome without the aid of a reference genome. Introduction As a result of the development of novel sequencing technologies, the years between 2008 and 2012 saw a large drop in the cost of sequencing. Per megabase and genome, the cost dropped to 1/100,000th and 1/10,000th of the price, respectively. Prior to this, only transcriptomes of organisms that were of broad interest and utility to scientific research were sequenced; however, these developed in 2010s high-throughput sequencing (also called next-generation sequencing) technologies are both cost- and labor- effective, and the range of organisms studied via these methods is expanding. Transcriptomes have subsequently been created for chickpea, planarians, '' Parhyale hawaiensis'', as well as the brains of the Nile crocodile, the corn snake, the bearded dragon, and the red-eared slider, to name just a few. Examining non-model organ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
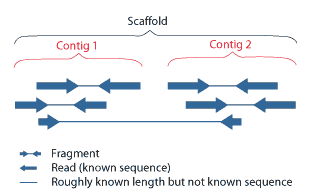
Contig
A contig (from ''contiguous'') is a set of overlapping DNA segments that together represent a consensus region of DNA.Gregory, S. ''Contig Assembly''. Encyclopedia of Life Sciences, 2005. In bottom-up sequencing projects, a contig refers to overlapping sequence data ( reads); in top-down sequencing projects, contig refers to the overlapping clones that form a physical map of the genome that is used to guide sequencing and assembly.Dear, P. H. ''Genome Mapping''. Encyclopedia of Life Sciences, 2005. . Contigs can thus refer both to overlapping DNA sequences and to overlapping physical segments (fragments) contained in clones depending on the context. Original definition of contig In 1980, Staden wrote: ''In order to make it easier to talk about our data gained by the shotgun method of sequencing we have invented the word "contig". A contig is a set of gel readings that are related to one another by overlap of their sequences. All gel readings belong to one and only one con ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Aerobic Organism
Aerobic means "requiring air," in which "air" usually means oxygen. Aerobic may also refer to * Aerobic exercise, prolonged exercise of moderate intensity * Aerobics, a form of aerobic exercise * Aerobic respiration, the aerobic process of cellular respiration * Aerobic organism Aerobic means "requiring air," in which "air" usually means oxygen. Aerobic may also refer to * Aerobic exercise, prolonged exercise of moderate intensity * Aerobics Aerobics is a form of physical exercise that combines rhythmic aerobic exe ..., a living thing with an oxygen-based metabolism See also * Anaerobic (other) {{disambiguation ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |