|
Locus Of Enterocyte Effacement
The locus of enterocyte effacement (LEE) is a moderately conserved pathogenicity island consisting of 35,000 base pairs in the bacteria ''Escherichia coli'' genome. The LEE encodes the Type III secretion system and associated chaperones and effector proteins responsible for attaching and effacing (AE) lesions in the large intestine. These proteins include intimin, Tir, EspC, EspF, EspH, and Map protein. The LEE has a 38% G+C ratio. See also *Locus of enterocyte effacement-encoded regulator The locus of enterocyte effacement-encoded regulator (Ler) is a regulatory protein that controls bacterial pathogenicity of enteropathogenic ''Escherichia coli'' (EPEC) and enterohemorrhagic ''Escherichia coli'' (EHEC). More specifically, Ler regu ... Cell biology {{microbiology-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pathogenicity Island
Pathogenicity islands (PAIs), as termed in 1990, are a distinct class of genomic islands acquired by microorganisms through horizontal gene transfer. Pathogenicity islands are found in both animal and plant pathogens. Additionally, PAIs are found in both gram-positive and gram-negative bacteria. They are transferred through horizontal gene transfer events such as transfer by a plasmid, phage, or conjugative transposon. Therefore, PAIs contribute to microorganisms' ability to evolve. One species of bacteria may have more than one PAI. For example, ''Salmonella'' has at least five. An analogous genomic structure in rhizobia is termed a '' symbiosis island''. Properties Pathogenicity islands (PAIs) are gene clusters incorporated in the genome, chromosomally or extrachromosomally, of pathogenic organisms, but are usually absent from those nonpathogenic organisms of the same or closely related species. They may be located on a bacterial chromosome or may be transferred within a plas ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Base Pairs
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA and RNA. Dictated by specific hydrogen bonding patterns, "Watson–Crick" (or "Watson–Crick–Franklin") base pairs (guanine–cytosine and adenine–thymine) allow the DNA helix to maintain a regular helical structure that is subtly dependent on its nucleotide sequence. The complementary nature of this based-paired structure provides a redundant copy of the genetic information encoded within each strand of DNA. The regular structure and data redundancy provided by the DNA double helix make DNA well suited to the storage of genetic information, while base-pairing between DNA and incoming nucleotides provides the mechanism through which DNA polymerase replicates DNA and RNA polymerase transcribes DNA into RNA. Many DNA-binding proteins ca ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Escherichia Coli
''Escherichia coli'' (),Wells, J. C. (2000) Longman Pronunciation Dictionary. Harlow ngland Pearson Education Ltd. also known as ''E. coli'' (), is a Gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus ''Escherichia'' that is commonly found in the lower intestine of warm-blooded organisms. Most ''E. coli'' strains are harmless, but some serotypes ( EPEC, ETEC etc.) can cause serious food poisoning in their hosts, and are occasionally responsible for food contamination incidents that prompt product recalls. Most strains do not cause disease in humans and are part of the normal microbiota of the gut; such strains are harmless or even beneficial to humans (although these strains tend to be less studied than the pathogenic ones). For example, some strains of ''E. coli'' benefit their hosts by producing vitamin K2 or by preventing the colonization of the intestine by pathogenic bacteria. These mutually beneficial relationships between ''E. col ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
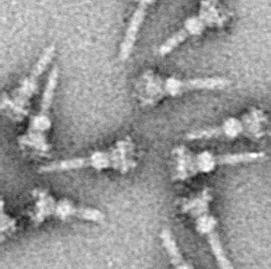
Type III Secretion System
The type III secretion system (T3SS or TTSS), also called the injectisome, is one of the bacterial secretion systems used by bacteria to secrete their effector proteins into the host's cells to promote virulence and colonisation. The T3SS is a needle-like protein complex found in several species of pathogenic gram-negative bacteria. Overview The term Type III secretion system was coined in 1993. This secretion system is distinguished from at least five other secretion systems found in gram-negative bacteria. Many animal and plant associated bacteria possess similar T3SSs. These T3SSs are similar as a result of divergent evolution and phylogenetic analysis supports a model in which gram-negative bacteria can transfer the T3SS gene cassette horizontally to other species. The most researched T3SSs are from species of ''Shigella'' (causes bacillary dysentery), ''Salmonella'' (typhoid fever), ''Escherichia coli'' (Gut flora, some strains cause food poisoning), ''Vibrio'' (gastr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Intimin
Intimin is a virulence factor ( adhesin) of EPEC (''e.g.'' ''E. coli'' O127:H6) and EHEC (''e.g. E. coli'' O157:H7) ''E. coli'' strains. It is an attaching and effacing (A/E) protein, which with other virulence factors is necessary and responsible for enteropathogenic and enterohaemorrhagic diarrhoea. Intimin is expressed on the bacterial cell surface where it can bind to its receptor Tir (Translocated intimin receptor). Tir, along with over 25 other bacterial proteins, is secreted from attaching and effacing ''E. coli'' directly into the cytoplasm of intestinal epithelial cells by a Type three secretion system. Once within the cytoplasm of the host cell, Tir is inserted into the plasma membrane, allowing surface exposure and intimin binding. Tir-intimin interaction mediates tight binding of enteropathogenic and enterohaemorrhagic ''E.coli'' to the intestinal epithelia, resulting in the formation of effacing lesions on intestinal epithelia. The structure of the C-terminal ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Map Protein
A map is a symbolic depiction emphasizing relationships between elements of some space, such as objects, regions, or themes. Many maps are static, fixed to paper or some other durable medium, while others are dynamic or interactive. Although most commonly used to depict geography, maps may represent any space, real or fictional, without regard to context or scale, such as in brain mapping, DNA mapping, or computer network topology mapping. The space being mapped may be two dimensional, such as the surface of the earth, three dimensional, such as the interior of the earth, or even more abstract spaces of any dimension, such as arise in modeling phenomena having many independent variables. Although the earliest maps known are of the heavens, geographic maps of territory have a very long tradition and exist from ancient times. The word "map" comes from the , wherein ''mappa'' meant 'napkin' or 'cloth' and ''mundi'' 'the world'. Thus, "map" became a shortened term referring to ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Locus Of Enterocyte Effacement-encoded Regulator
The locus of enterocyte effacement-encoded regulator (Ler) is a regulatory protein that controls bacterial pathogenicity of enteropathogenic ''Escherichia coli'' (EPEC) and enterohemorrhagic ''Escherichia coli'' (EHEC). More specifically, Ler regulates the locus of enterocyte effacement (LEE) pathogenicity island genes, which are responsible for creating intestinal attachment and effacing lesions and subsequent diarrhea: LEE1, LEE2, and LEE3. LEE1, 2, and 3 carry the information necessary for a type III secretion system. The transcript encoding the Ler protein is the open reading frame 1 on the LEE1 operon. The mechanism of Ler regulation involves competition with histone-like nucleoid structuring protein (H-NS), a negative regulator of the LEE pathogenicity island. Ler is regulated by many factors such as plasmid encoded regulator (Per), integration host factor, Fis, BipA, a positive regulatory loop involving GrlA, and quorum sensing mediated by ''luxS''. __TOC__ Mechanis ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |