|
Intrinsically Unstructured Protein
In molecular biology, an intrinsically disordered protein (IDP) is a protein that lacks a fixed or ordered three-dimensional structure, typically in the absence of its macromolecular interaction partners, such as other proteins or RNA. IDPs range from fully unstructured to partially structured and include random coil, molten globule-like aggregates, or flexible linkers in large multi-domain proteins. They are sometimes considered as a separate class of proteins along with globular, fibrous and membrane proteins. IDPs are a very large and functionally important class of proteins and their discovery has disproved the idea that three-dimensional structures of proteins must be fixed to accomplish their biological functions. For example, IDPs have been identified to participate in weak multivalent interactions that are highly cooperative and dynamic, lending them importance in DNA regulation and in cell signaling. Many IDPs can also adopt a fixed three-dimensional structu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
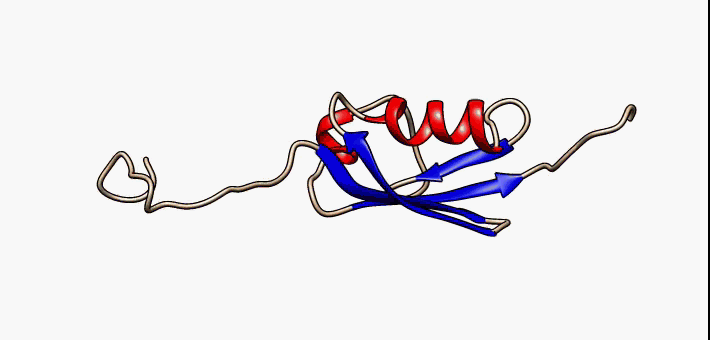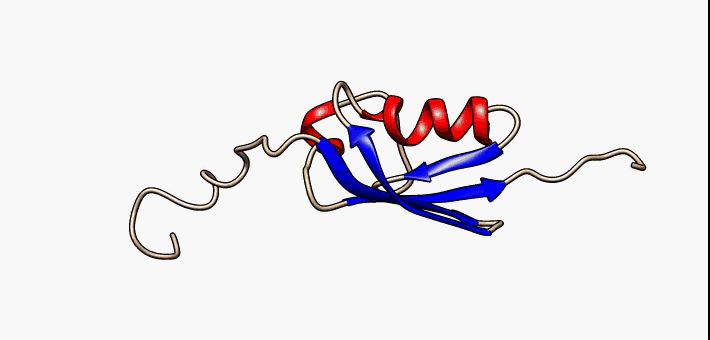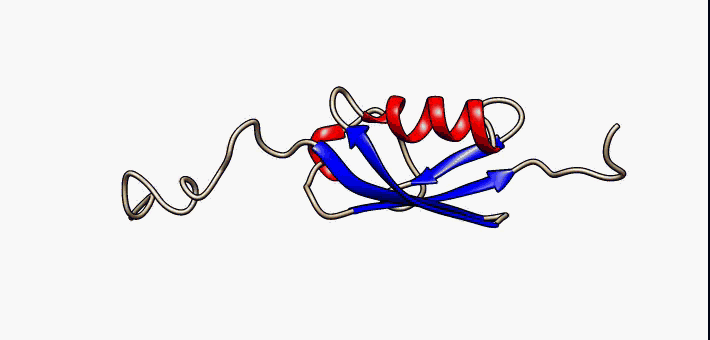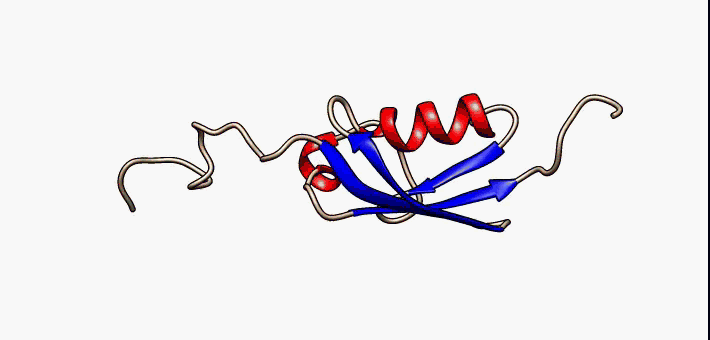
PDB 2fft EBI
PDB may refer to: * Chess Problem Database Server (PDB Server) * 1,4-Dichlorobenzene (paradichlorobenzene) * Party of German-speaking Belgians, (German: '), a political party and predecessor of the ProDG * PDB (Palm OS), a container format for record databases in Palm OS, Garnet OS and Access Linux Platform * ''Pee Dee Belemnite'', a standard for stable Carbon-13 and Oxygen-18 isotopes; see * Pluggable database, such as an Oracle Database in a multitenancy environment * Potato dextrose broth, a common microbiological growth media * Pousette-Dart Band * President's Daily Brief or Briefing or Bulletin, a top-secret intelligence document produced each morning for the U.S. President * Program database, a file format for storing debugging information * Promised Day Brigade, an Iraqi Shia organisation * Protein Data Bank * Protein Data Bank (file format) * Python Debugger, see Python (programming language) Python is a high-level, general-purpose programming language. Its design philo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Proteome
The proteome is the entire set of proteins that is, or can be, expressed by a genome, cell, tissue, or organism at a certain time. It is the set of expressed proteins in a given type of cell or organism, at a given time, under defined conditions. Proteomics is the study of the proteome. Types of proteomes While proteome generally refers to the proteome of an organism, multicellular organisms may have very different proteomes in different cells, hence it is important to distinguish proteomes in cells and organisms. A cellular proteome is the collection of proteins found in a particular cell type under a particular set of environmental conditions such as exposure to hormone stimulation. It can also be useful to consider an organism's complete proteome, which can be conceptualized as the complete set of proteins from all of the various cellular proteomes. This is very roughly the protein equivalent of the genome. The term ''proteome'' has also been used to refer to the collectio ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genome
In the fields of molecular biology and genetics, a genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as regulatory sequences (see non-coding DNA), and often a substantial fraction of 'junk' DNA with no evident function. Almost all eukaryotes have mitochondria and a small mitochondrial genome. Algae and plants also contain chloroplasts with a chloroplast genome. The study of the genome is called genomics. The genomes of many organisms have been sequenced and various regions have been annotated. The International Human Genome Project reported the sequence of the genome for ''Homo sapiens'' in 200The Human Genome Project although the initial "finished" sequence was missing 8% of the genome consisting mostly of repetitive sequences. With advancements in technology that could handle sequenci ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bioinformatic
Bioinformatics () is an interdisciplinary field that develops methods and software tools for understanding biological data, in particular when the data sets are large and complex. As an interdisciplinary field of science, bioinformatics combines biology, chemistry, physics, computer science, information engineering, mathematics and statistics to analyze and interpret the biological data. Bioinformatics has been used for ''in silico'' analyses of biological queries using computational and statistical techniques. Bioinformatics includes biological studies that use computer programming as part of their methodology, as well as specific analysis "pipelines" that are repeatedly used, particularly in the field of genomics. Common uses of bioinformatics include the identification of candidates genes and single nucleotide polymorphisms (SNPs). Often, such identification is made with the aim to better understand the genetic basis of disease, unique adaptations, desirable properties (esp. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tau Protein
The tau proteins (abbreviated from tubulin associated unit) are a group of six highly soluble protein isoforms produced by alternative splicing from the gene ''MAPT'' (microtubule-associated protein tau). They have roles primarily in maintaining the stability of microtubules in axons and are abundant in the neurons of the central nervous system (CNS), where the cerebral cortex has the highest abundance. They are less common elsewhere but are also expressed at very low levels in CNS astrocytes and oligodendrocytes. Pathologies and dementias of the nervous system such as Alzheimer's disease and Parkinson's disease are associated with tau proteins that have become hyperphosphorylated insoluble aggregates called neurofibrillary tangles. The tau proteins were identified in 1975 as heat-stable proteins essential for microtubule assembly, and since then they have been characterized as intrinsically disordered proteins. Function Microtubule stabilization Tau proteins are found mo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alpha-synuclein
Alpha-synuclein is a protein that, in humans, is encoded by the ''SNCA'' gene. Alpha-synuclein is a neuronal protein that regulates synaptic vesicle trafficking and subsequent neurotransmitter release. It is abundant in the brain, while smaller amounts are found in the heart, muscle and other tissues. In the brain, alpha-synuclein is found mainly in the axon terminals of presynaptic neurons. Within these terminals, alpha-synuclein interacts with phospholipids and proteins. Presynaptic terminals release chemical messengers, called neurotransmitters, from compartments known as synaptic vesicles. The release of neurotransmitters relays signals between neurons and is critical for normal brain function. The human alpha-synuclein protein is made of 140 amino acids. An alpha-synuclein fragment, known as the non- Abeta component (NAC) of Alzheimer's disease amyloid, originally found in an amyloid-enriched fraction, was shown to be a fragment of its precursor protein, NACP. It was later de ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nuclear Magnetic Resonance Spectroscopy Of Proteins
Nuclear magnetic resonance spectroscopy of proteins (usually abbreviated protein NMR) is a field of structural biology in which NMR spectroscopy is used to obtain information about the structure and dynamics of proteins, and also nucleic acids, and their complexes. The field was pioneered by Richard R. Ernst and Kurt Wüthrich at the ETH, and by Ad Bax, Marius Clore, Angela Gronenborn at the NIH, and Gerhard Wagner at Harvard University, among others. Structure determination by NMR spectroscopy usually consists of several phases, each using a separate set of highly specialized techniques. The sample is prepared, measurements are made, interpretive approaches are applied, and a structure is calculated and validated. NMR involves the quantum-mechanical properties of the central core ("nucleus") of the atom. These properties depend on the local molecular environment, and their measurement provides a map of how the atoms are linked chemically, how close they are in space, and how rapid ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Electron Density
In quantum chemistry, electron density or electronic density is the measure of the probability of an electron being present at an infinitesimal element of space surrounding any given point. It is a scalar quantity depending upon three spatial variables and is typically denoted as either \rho(\textbf r) or n(\textbf r). The density is determined, through definition, by the normalised N-electron wavefunction which itself depends upon 4N variables (3N spatial and N spin coordinates). Conversely, the density determines the wave function modulo up to a phase factor, providing the formal foundation of density functional theory. According to quantum mechanics, due to the uncertainty principle on an atomic scale the exact location of an electron cannot be predicted, only the probability of its being at a given position; therefore electrons in atoms and molecules act as if they are "smeared out" in space. For one-electron systems, the electron density at any point is proportional to th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Anfinsen's Dogma
Anfinsen's dogma, also known as the thermodynamic hypothesis, is a postulate in molecular biology. It states that, at least for a small globular protein in its standard physiological environment, the native structure is determined only by the protein's amino acid sequence. The dogma was championed by the Nobel Prize Laureate Christian B. Anfinsen from his research on the folding of ribonuclease A. The postulate amounts to saying that, at the environmental conditions (temperature, solvent concentration and composition, etc.) at which folding occurs, the native structure is a unique, stable and kinetically accessible minimum of the free energy. In other words, there are three conditions for formation of a unique protein structure: *Uniqueness – Requires that the sequence does not have any other configuration with a comparable free energy. Hence the free energy minimum must be ''unchallenged''. *Stability – Small changes in the surrounding environment cannot give rise to changes ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Levinthal's Paradox
Levinthal's paradox is a thought experiment, also constituting a self-reference in the theory of protein folding. In 1969, Cyrus Levinthal noted that, because of the very large number of degrees of freedom in an unfolded polypeptide chain, the molecule has an astronomical number of possible conformations. An estimate of 10300 was made in one of his papers (often incorrectly cited as the 1968 paper). For example, a polypeptide of 100 residues will have 99 peptide bonds, and therefore 198 different phi and psi bond angles. If each of these bond angles can be in one of three stable conformations, the protein may misfold into a maximum of 3198 different conformations (including any possible folding redundancy). Therefore, if a protein were to attain its correctly folded configuration by sequentially sampling all the possible conformations, it would require a time longer than the age of the universe to arrive at its correct native conformation. This is true even if conformations are ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Central Dogma Of Molecular Biology
The central dogma of molecular biology is an explanation of the flow of genetic information within a biological system. It is often stated as "DNA makes RNA, and RNA makes protein", although this is not its original meaning. It was first stated by Francis Crick in 1957, then published in 1958: He re-stated it in a ''Nature'' paper published in 1970: "The central dogma of molecular biology deals with the detailed residue-by-residue transfer of sequential information. It states that such information cannot be transferred back from protein to either protein or nucleic acid." A second version of the central dogma is popular but incorrect. This is the simplistic DNA → RNA → protein pathway published by James Watson in the first edition of ''The Molecular Biology of the Gene'' (1965). Watson's version differs from Crick's because Watson describes a two-step (DNA → RNA and RNA → protein) process as the central dogma. While the dogma, as originally stated by Crick, remains ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |