|
Integrases
Retroviral integrase (IN) is an enzyme produced by a retrovirus (such as HIV) that integrates—forms covalent links between—its genetic information into that of the host cell it infects. Retroviral INs are not to be confused with phage integrases (recombinases) used in biotechnology, such as λ phage integrase, as discussed in site-specific recombination. The macromolecular complex of an IN macromolecule bound to the ends of the viral DNA ends has been referred to as the '' intasome''; IN is a key component in this and the retroviral pre-integration complex. Structure All retroviral IN proteins contain three canonical domains, connected by flexible linkers: ● an N-terminal HH-CC zinc-binding domain (a three-helical bundle stabilized by coordination of a Zn(II) cation) ● a catalytic core domain (RNaseH fold) ● a C-terminal DNA-binding domain ( SH3 fold). Crystal and NMR structures of the individual domains and 2-domain constructs of integrases fro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Lambda Phage
''Enterobacteria phage λ'' (lambda phage, coliphage λ, officially ''Escherichia virus Lambda'') is a bacterial virus, or bacteriophage, that infects the bacterial species ''Escherichia coli'' (''E. coli''). It was discovered by Esther Lederberg in 1950. The wild type of this virus has a temperate life cycle that allows it to either reside within the genome of its host through lysogeny or enter into a lytic phase, during which it kills and lyses the cell to produce offspring. Lambda strains, mutated at specific sites, are unable to lysogenize cells; instead, they grow and enter the lytic cycle after superinfecting an already lysogenized cell. The phage particle consists of a head (also known as a capsid), a tail, and tail fibers (see image of virus below). The head contains the phage's double-strand linear DNA genome. During infection, the phage particle recognizes and binds to its host, ''E. coli'', causing DNA in the head of the phage to be ejected through the tail into the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
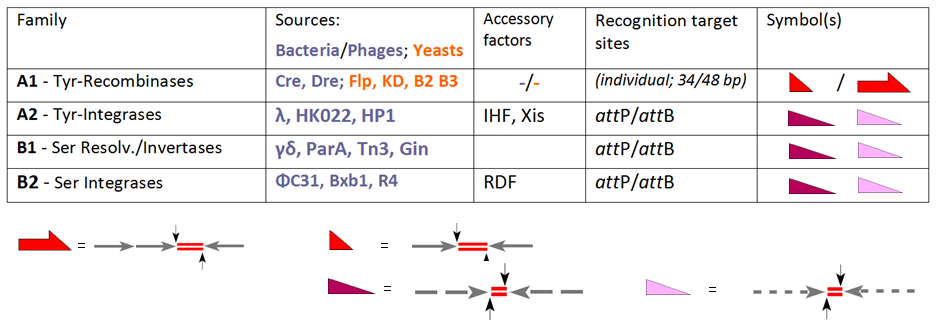
Site-specific Recombination
Site-specific recombination, also known as conservative site-specific recombination, is a type of genetic recombination in which DNA strand exchange takes place between segments possessing at least a certain degree of sequence homology. Enzymes known as site-specific recombinases (SSRs) perform rearrangements of DNA segments by recognizing and binding to short, specific DNA sequences (sites), at which they cleave the DNA backbone, exchange the two DNA helices involved, and rejoin the DNA strands. In some cases the presence of a recombinase enzyme and the recombination sites is sufficient for the reaction to proceed; in other systems a number of accessory proteins and/or accessory sites are required. Many different genome modification strategies, among these recombinase-mediated cassette exchange (RMCE), an advanced approach for the targeted introduction of transcription units into predetermined genomic loci, rely on SSRs. Site-specific recombination systems are highly specific, fa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phage Integrase
Site-specific recombination, also known as conservative site-specific recombination, is a type of genetic recombination in which DNA strand exchange takes place between segments possessing at least a certain degree of sequence homology. Enzymes known as site-specific recombinases (SSRs) perform rearrangements of DNA segments by recognizing and binding to short, specific DNA sequences (sites), at which they cleave the DNA backbone, exchange the two DNA helices involved, and rejoin the DNA strands. In some cases the presence of a recombinase enzyme and the recombination sites is sufficient for the reaction to proceed; in other systems a number of accessory proteins and/or accessory sites are required. Many different genome modification strategies, among these recombinase-mediated cassette exchange (RMCE), an advanced approach for the targeted introduction of transcription units into predetermined genomic loci, rely on SSRs. Site-specific recombination systems are highly specific, fa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Site-specific Recombinase Technology
Site-specific recombinase technologies are genome engineering tools that depend on recombinase enzymes to replace targeted sections of DNA. History In the late 1980s gene targeting in murine embryonic stem cells (ESCs) enabled the transmission of mutations into the mouse germ line, and emerged as a novel option to study the genetic basis of regulatory networks as they exist in the genome. Still, classical gene targeting proved to be limited in several ways as gene functions became irreversibly destroyed by the marker gene that had to be introduced for selecting recombinant ESCs. These early steps led to animals in which the mutation was present in all cells of the body from the beginning leading to complex phenotypes and/or early lethality. There was a clear need for methods to restrict these mutations to specific points in development and specific cell types. This dream became reality when groups in the USA were able to introduce bacteriophage and yeast-derived site-specific rec ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Site Specific Recombination
Site-specific recombination, also known as conservative site-specific recombination, is a type of genetic recombination in which DNA strand exchange takes place between segments possessing at least a certain degree of sequence homology. Enzymes known as site-specific recombinases (SSRs) perform rearrangements of DNA segments by recognizing and binding to short, specific DNA sequences (sites), at which they cleave the DNA backbone, exchange the two DNA helices involved, and rejoin the DNA strands. In some cases the presence of a recombinase enzyme and the recombination sites is sufficient for the reaction to proceed; in other systems a number of accessory proteins and/or accessory sites are required. Many different genome modification strategies, among these recombinase-mediated cassette exchange (RMCE), an advanced approach for the targeted introduction of transcription units into predetermined genomic loci, rely on SSRs. Site-specific recombination systems are highly specific, fa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Human Foamy Virus
Human foamy virus (HFV) is a retrovirus and specifically belongs to the genus ''Spumavirus''. The spumaviruses are complex and significantly different from the other six genera of retroviruses in several ways. The foamy viruses derive their name from the characteristic ‘foamy’ appearance of the cytopathic effect (CPE) induced in the cells. Foamy virus in humans occurs only as a result of zoonotic infection. Discovery The first description of foamy virus (FV) was in 1954. It was found as a contaminant in primary monkey kidney cultures. The first isolate of the “foamy viral agent” was in 1955. Not too long after this, it was isolated from a wide variety of New and Old World monkeys, cats, and cows. It was not until several years later that humans entered the scene. In 1971, a viral agent with FV-like characteristics was isolated from lymphoblastoid cells released from a human nasopharyngeal carcinoma (NPC) from a Kenyan patient. The agent was termed a human FV bec ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Reverse Transcriptase
A reverse transcriptase (RT) is an enzyme used to generate complementary DNA (cDNA) from an RNA template, a process termed reverse transcription. Reverse transcriptases are used by viruses such as HIV and hepatitis B to replicate their genomes, by retrotransposon mobile genetic elements to proliferate within the host genome, and by eukaryotic cells to extend the telomeres at the ends of their linear chromosomes. Contrary to a widely held belief, the process does not violate the flows of genetic information as described by the classical central dogma, as transfers of information from RNA to DNA are explicitly held possible. Retroviral RT has three sequential biochemical activities: RNA-dependent DNA polymerase activity, ribonuclease H (RNase H), and DNA-dependent DNA polymerase activity. Collectively, these activities enable the enzyme to convert single-stranded RNA into double-stranded cDNA. In retroviruses and retrotransposons, this cDNA can then integrate into the host genom ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transesterification
In organic chemistry, transesterification is the process of exchanging the organic group R″ of an ester with the organic group R' of an alcohol. These reactions are often catalyzed by the addition of an acid or base catalyst. The reaction can also be accomplished with the help of other enzymes, particularly lipases (one example is the lipase E.C.3.1.1.3). Strong acids catalyse the reaction by donating a proton to the carbonyl group, thus making it a more potent electrophile, whereas bases catalyse the reaction by removing a proton from the alcohol, thus making it more nucleophilic. If the alcohol produced by the reaction can be separated from the reactants by distillation this will drive the equilibrium toward the products, this means that esters with larger alkoxy groups can be made from methyl or ethyl esters in high purity by heating the mixture of ester, acid/base, and large alcohol. Mechanism In the transesterification mechanism, the carbonyl carbon of the starting e ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Covalent
A covalent bond is a chemical bond that involves the sharing of electrons to form electron pairs between atoms. These electron pairs are known as shared pairs or bonding pairs. The stable balance of attractive and repulsive forces between atoms, when they share electrons, is known as covalent bonding. For many molecules, the sharing of electrons allows each atom to attain the equivalent of a full valence shell, corresponding to a stable electronic configuration. In organic chemistry, covalent bonding is much more common than ionic bonding. Covalent bonding also includes many kinds of interactions, including σ-bonding, π-bonding, metal-to-metal bonding, agostic interactions, bent bonds, three-center two-electron bonds and three-center four-electron bonds. The term ''covalent bond'' dates from 1939. The prefix ''co-'' means ''jointly, associated in action, partnered to a lesser degree, '' etc.; thus a "co-valent bond", in essence, means that the atoms share " valence", such a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are catalytic RNA molecules, called ribozymes. Enzymes' specificity comes from their unique three-dimensional structures. Like all catalysts, enzymes increase the reaction ra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
The Actual HIV Integrase
''The'' () is a grammatical article in English, denoting persons or things already mentioned, under discussion, implied or otherwise presumed familiar to listeners, readers, or speakers. It is the definite article in English. ''The'' is the most frequently used word in the English language; studies and analyses of texts have found it to account for seven percent of all printed English-language words. It is derived from gendered articles in Old English which combined in Middle English and now has a single form used with pronouns of any gender. The word can be used with both singular and plural nouns, and with a noun that starts with any letter. This is different from many other languages, which have different forms of the definite article for different genders or numbers. Pronunciation In most dialects, "the" is pronounced as (with the voiced dental fricative followed by a schwa) when followed by a consonant sound, and as (homophone of pronoun ''thee'') when followed by a v ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tetrameric Protein
A tetrameric protein is a protein with a quaternary structure of four subunits (tetrameric). Homotetramers have four identical subunits (such as glutathione S-transferase), and heterotetramers are complexes of different subunits. A tetramer can be assembled as dimer of dimers with two homodimer subunits (such as sorbitol dehydrogenase), or two heterodimer subunits (such as hemoglobin). Subunit interactions in tetramers The interactions between subunits forming a tetramer is primarily determined by non covalent interaction. Hydrophobic effects, hydrogen bonds and electrostatic interactions are the primary sources for this binding process between subunits. For homotetrameric proteins such as Sorbitol dehydrogenase (SDH), the structure is believed to have evolved going from a monomeric to a dimeric and finally a tetrameric structure in evolution. The binding process in SDH and many other tetrameric enzymes can be described by the gain in free energy which can be determined from ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |




.png)
