|
Histone-modifying Enzymes
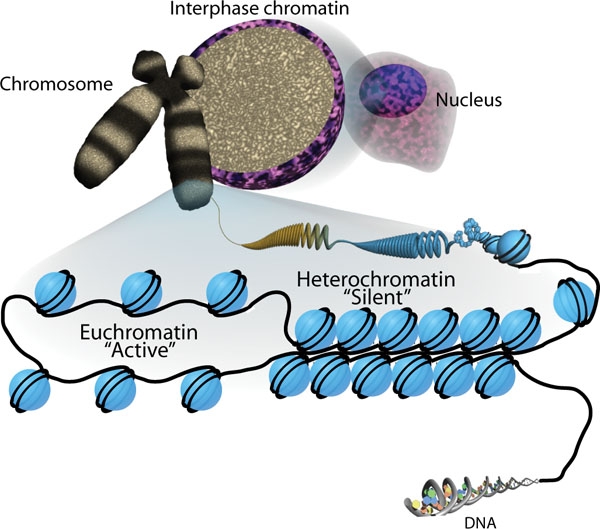
Histone-modifying enzymes are enzymes involved in the modification of histone substrates after protein translation and affect cellular processes including gene expression. To safely store the eukaryotic genome, DNA is wrapped around four core histone proteins (H3, H4, H2A, H2B), which then join to form nucleosomes. These nucleosomes further fold together into highly condensed chromatin, which renders the organism's genetic material far less accessible to the factors required for gene transcription, DNA replication, recombination and repair. Subsequently, eukaryotic organisms have developed intricate mechanisms to overcome this repressive barrier imposed by the chromatin through histone modification, a type of post-translational modification which typically involves covalently attaching certain groups to histone residues. Once added to the histone, these groups (directly or indirectly) elicit either a loose and open histone conformation, euchromatin, or a tight and closed histone ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
The Basic Unit Of Chromatin Organization Is The Nucleosome, Which Comprises 147 Bp Of DNA Wrapped Ar
''The'' () is a grammatical article in English, denoting persons or things already mentioned, under discussion, implied or otherwise presumed familiar to listeners, readers, or speakers. It is the definite article in English. ''The'' is the most frequently used word in the English language; studies and analyses of texts have found it to account for seven percent of all printed English-language words. It is derived from gendered articles in Old English which combined in Middle English and now has a single form used with pronouns of any gender. The word can be used with both singular and plural nouns, and with a noun that starts with any letter. This is different from many other languages, which have different forms of the definite article for different genders or numbers. Pronunciation In most dialects, "the" is pronounced as (with the voiced dental fricative followed by a schwa) when followed by a consonant sound, and as (homophone of pronoun ''thee'') when followed by a v ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Covalent Bond
A covalent bond is a chemical bond that involves the sharing of electrons to form electron pairs between atoms. These electron pairs are known as shared pairs or bonding pairs. The stable balance of attractive and repulsive forces between atoms, when they share electrons, is known as covalent bonding. For many molecules, the sharing of electrons allows each atom to attain the equivalent of a full valence shell, corresponding to a stable electronic configuration. In organic chemistry, covalent bonding is much more common than ionic bonding. Covalent bonding also includes many kinds of interactions, including σ-bonding, π-bonding, metal-to-metal bonding, agostic interactions, bent bonds, three-center two-electron bonds and three-center four-electron bonds. The term ''covalent bond'' dates from 1939. The prefix ''co-'' means ''jointly, associated in action, partnered to a lesser degree, '' etc.; thus a "co-valent bond", in essence, means that the atoms share " valence", such a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Proline Isomerization
In epigenetics, proline isomerization is the effect that ''cis-trans'' isomerization of the amino acid proline has on the regulation of gene expression. Similar to aspartic acid, the amino acid proline has the rare property of being able to occupy both ''cis'' and ''trans'' isomers of its prolyl peptide bonds with ease. Peptidyl-prolyl isomerase, or PPIase, is an enzyme very commonly associated with proline isomerization due to their ability to catalyze the isomerization of prolines. PPIases are present in three types: cyclophilins, FK507-binding proteins, and the parvulins. PPIase enzymes catalyze the transition of proline between ''cis'' and ''trans'' isomers and are essential to the numerous biological functions controlled and affected by prolyl isomerization (i.e. cell signalling, protein folding, and epigenetic modifications) Without PPIases, prolyl peptide bonds will slowly switch between ''cis'' and ''trans'' isomers, a process that can lock proteins in a nonnative structure ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Citrullination
Citrullination or deimination is the conversion of the amino acid arginine in a protein into the amino acid citrulline. Citrulline is not one of the 20 standard amino acids encoded by DNA in the genetic code. Instead, it is the result of a post-translational modification. Citrullination is distinct from the formation of the free amino acid citrulline as part of the urea cycle or as a byproduct of enzymes of the nitric oxide synthase family. Enzymes called arginine deiminases (ADIs) catalyze the deimination of free arginine, while protein arginine deiminases or peptidylarginine deiminases (PADs) replace the primary ketimine group (>C=NH) by a ketone group (>C=O). Arginine is positively charged at a neutral pH, whereas citrulline has no net charge. This increases the hydrophobicity of the protein, which can lead to changes in protein folding, affecting the structure and function. The immune system can attack citrullinated proteins, leading to autoimmune diseases such as rheumatoid ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ADP-ribosylation
ADP-ribosylation is the addition of one or more ADP-ribose moieties to a protein. It is a reversible post-translational modification that is involved in many cellular processes, including cell signaling, DNA repair, gene regulation and apoptosis. Improper ADP-ribosylation has been implicated in some forms of cancer. It is also the basis for the toxicity of bacterial compounds such as cholera toxin, diphtheria toxin, and others. History The first suggestion of ADP-ribosylation surfaced during the early 1960s. At this time, Pierre Chambon and coworkers observed the incorporation of ATP into hen liver nuclei extract. After extensive studies on the acid insoluble fraction, several different research laboratories were able to identify ADP-ribose, derived from NAD+, as the incorporated group. Several years later, the enzymes responsible for this incorporation were identified and given the name poly(ADP-ribose)polymerase. Originally, this group was thought to be a linear sequence of AD ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SUMO Protein
In molecular biology, SUMO (Small Ubiquitin-like Modifier) proteins are a family of small proteins that are covalently attached to and detached from other proteins in cells to modify their function. This process is called SUMOylation (sometimes written sumoylation). SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle. SUMO proteins are similar to ubiquitin and are considered members of the ubiquitin-like protein family. SUMOylation is directed by an enzymatic cascade analogous to that involved in ubiquitination. In contrast to ubiquitin, SUMO is not used to tag proteins for degradation. Mature SUMO is produced when the last four amino acids of the C-terminus have been cleaved off to allow formation of an isopeptide bond between the C-terminal glycine residue of SUMO and an acceptor lysine on ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
O-GlcNAc
''O''-GlcNAc (short for ''O''-linked GlcNAc or ''O''-linked β-''N''-acetylglucosamine) is a reversible enzymatic post-translational modification that is found on serine and threonine residues of nucleocytoplasmic proteins. The modification is characterized by a β-glycosidic bond between the hydroxyl group of serine or threonine side chains and ''N''-acetylglucosamine (GlcNAc). ''O''-GlcNAc differs from other forms of protein glycosylation: (i) ''O''-GlcNAc is not elongated or modified to form more complex glycan structures, (ii) ''O''-GlcNAc is almost exclusively found on nuclear and cytoplasmic proteins rather than membrane proteins and secretory proteins, and (iii) ''O''-GlcNAc is a highly dynamic modification that turns over more rapidly than the proteins which it modifies. ''O''-GlcNAc is conserved across metazoans. Due to the dynamic nature of ''O''-GlcNAc and its presence on serine and threonine residues, ''O''-GlcNAcylation is similar to protein phosphorylation in some r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Functional Group
In organic chemistry, a functional group is a substituent or moiety in a molecule that causes the molecule's characteristic chemical reactions. The same functional group will undergo the same or similar chemical reactions regardless of the rest of the molecule's composition. This enables systematic prediction of chemical reactions and behavior of chemical compounds and the design of chemical synthesis. The reactivity of a functional group can be modified by other functional groups nearby. Functional group interconversion can be used in retrosynthetic analysis to plan organic synthesis. A functional group is a group of atoms in a molecule with distinctive chemical properties, regardless of the other atoms in the molecule. The atoms in a functional group are linked to each other and to the rest of the molecule by covalent bonds. For repeating units of polymers, functional groups attach to their nonpolar core of carbon atoms and thus add chemical character to carbon chains. Fun ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ubiquitination
Ubiquitin is a small (8.6 kDa) regulatory protein found in most tissues of eukaryotic organisms, i.e., it is found ''ubiquitously''. It was discovered in 1975 by Gideon Goldstein and further characterized throughout the late 1970s and 1980s. Four genes in the human genome code for ubiquitin: UBB, UBC, UBA52 and RPS27A. The addition of ubiquitin to a substrate protein is called ubiquitylation (or, alternatively, ubiquitination or ubiquitinylation). Ubiquitylation affects proteins in many ways: it can mark them for degradation via the proteasome, alter their cellular location, affect their activity, and promote or prevent protein interactions. Ubiquitylation involves three main steps: activation, conjugation, and ligation, performed by ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), and ubiquitin ligases (E3s), respectively. The result of this sequential cascade is to bind ubiquitin to lysine residues on the protein substrate via an isopeptide bond, cy ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Phosphorylation
Protein phosphorylation is a reversible post-translational modification of proteins in which an amino acid residue is phosphorylated by a protein kinase by the addition of a covalently bound phosphate group. Phosphorylation alters the structural conformation of a protein, causing it to become either activated or deactivated, or otherwise modifying its function. Approximately 13000 human proteins have sites that are phosphorylated. The reverse reaction of phosphorylation is called dephosphorylation, and is catalyzed by protein phosphatases. Protein kinases and phosphatases work independently and in a balance to regulate the function of proteins. The amino acids most commonly phosphorylated are serine, threonine, tyrosine in eukaryotes, and also histidine in prokaryotes and plants (though it is now known to be common in humans). These phosphorylations play important and well-characterized roles in signaling pathways and metabolism. However, other amino acids can also be phosphory ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone Methylation
Histone methylation is a process by which methyl groups are transferred to amino acids of histone proteins that make up nucleosomes, which the DNA double helix wraps around to form chromosomes. Methylation of histones can either increase or decrease transcription of genes, depending on which amino acids in the histones are methylated, and how many methyl groups are attached. Methylation events that weaken chemical attractions between histone tails and DNA increase transcription because they enable the DNA to uncoil from nucleosomes so that transcription factor proteins and RNA polymerase can access the DNA. This process is critical for the regulation of gene expression that allows different cells to express different genes. Function Histone methylation, as a mechanism for modifying chromatin structure is associated with stimulation of neural pathways known to be important for formation of long-term memories and learning. Animal models have shown methylation and other epigenetic re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone Acetylation And Deacetylation
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei. They act as spools around which DNA winds to create structural units called nucleosomes. Nucleosomes in turn are wrapped into 30- nanometer fibers that form tightly packed chromatin. Histones prevent DNA from becoming tangled and protect it from DNA damage. In addition, histones play important roles in gene regulation and DNA replication. Without histones, unwound DNA in chromosomes would be very long. For example, each human cell has about 1.8 meters of DNA if completely stretched out; however, when wound about histones, this length is reduced to about 90 micrometers (0.09 mm) of 30 nm diameter chromatin fibers. There are five families of histones which are designated H1/H5 (linker histones), H2, H3, and H4 (core histones). The nucleosome core is formed of two H2A-H2B dimers and a H3-H4 tetramer. The tight wrapping of DNA around hist ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
.png)




