|
Hfq
The Hfq protein (also known as HF-I protein) encoded by the ''hfq'' gene was discovered in 1968 as an ''Escherichia coli'' host factor that was essential for replication of the bacteriophage Qβ. It is now clear that Hfq is an abundant bacterial RNA binding protein which has many important physiological roles that are usually mediated by interacting with Hfq binding sRNA. In ''E. coli'', Hfq mutants show multiple stress response related phenotypes. The Hfq protein is now known to regulate the translation of two major stress transcription factors ( σS (RpoS) and σE (RpoE) ) in Enterobacteria. It also regulates sRNA in ''Vibrio cholerae'', a specific example being MicX sRNA. In ''Salmonella typhimurium'', Hfq has been shown to be an essential virulence factor as its deletion attenuates the ability of ''S.typhimurium'' to invade epithelial cells, secrete virulence factors or survive in cultured macrophages. In ''Salmonella'', Hfq deletion mutants are also non motile and exhibit chro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hfq Binding SRNA
An Hfq binding sRNA is an sRNA that binds the bacterial RNA binding protein called Hfq. A number of bacterial small RNAs which have been shown to bind to Hfq have been characterised (see list). Many of these RNAs share a similar structure composed of three stem-loops. Several studies have expanded this list, and experimentally validated a total of 64 Hfq binding sRNA in ''Salmonella Typhimurium''. A transcriptome wide study on Hfq binding sites in ''Salmonella'' mapped 126 Hfq binding sites within sRNAs. Genomic SELEX has been used to show that Hfq binding RNAs are enriched in the sequence motif 5′-AAYAAYAA-3′. Genome-wide study identified 40 candidate Hfq-dependent sRNAs in plant pathogen ''Erwinia amylovora.'' 12 of them were confirmed by Northern blot. Bacterial Hfq binding sRNAs * DicF RNA * DsrA RNA * FnrS RNA * GadY RNA * GcvB RNA * IsrJ RNA * MicA RNA / SraD RNA * MicC RNA * MicF RNA * OmrA RNA / OmrB RNA / RygA RNA / RygB RNA / SraE RNA * OxyS RNA * Qrr RN ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
LSm 1I5L 1
In molecular biology, LSm proteins are a family of RNA-binding proteins found in virtually every cellular organism. LSm is a contraction of 'like Sm', because the first identified members of the LSm protein family were the Sm proteins. LSm proteins are defined by a characteristic three-dimensional structure and their assembly into rings of six or seven individual LSm protein molecules, and play a large number of various roles in mRNA processing and regulation. The Sm proteins were first discovered as antigens targeted by so-called anti-Sm antibodies in a patient with a form of systemic lupus erythematosus (SLE), a debilitating autoimmune disease. They were named Sm proteins in honor of Stephanie Smith, a patient who suffered from SLE. Other proteins with very similar structures were subsequently discovered and named LSm proteins. New members of the LSm protein family continue to be identified and reported. Proteins with similar structures are grouped into a hierarchy of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MicX SRNA
MicX sRNA (formerly known as A10) is a small non-coding RNA found in ''Vibrio cholerae''. It was given the name MicX as it has a similar function to MicA, MicC and MicF in '' E. coli''. MicX sRNA negatively regulates an outer membrane protein (coded for bVC0972 and also a component of an ABC transporter (genVC0620. These interactions were predicted and then confirmed using a DNA microarray. MicX was identified through a bioinformatics screen of ''V. cholerae'' having been previously predicted. Levels of transcription of this sRNA were compared under several conditions: it was found to be expressed on all tested mediums; richer mediums slightly reduced transcription; repression of certain sigma factors (δS and δE) did not change transcription but it was dramatically reduced in the absence of Hfq protein. This observation is in accordance with other sRNA expression patterns. The MicX RNA gene overlaps witVCA0943- a gene coding for a maltose transporter permease - but the ri ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA-OUT
RNA-OUT is a non-coding RNA that is antisense to the RNA-IN non-coding RNA. Transposition of insertion sequence IS10 is regulated by an anti-sense RNA which inhibits transposase expression when IS10 is present in multiple copies per cell. IS10 antisense pairing is facilitated by the RNA-binding protein, Hfq. RNA-OUT consists of a stem-loop domain topped by a flexibly paired loop; the 5′ end of the target molecule, RNA-IN, is complementary to the top of the loop, and complementarity extends for 35 nucleotides Nucleotides are organic molecules consisting of a nucleoside and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of which are essential biomolecules w ... down one side of RNA-OUT. References External links * Non-coding RNA {{molecular-cell-biology-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Photorhabdus Luminescens
''Photorhabdus luminescens'' (previously called ''Xenorhabdus luminescens'') is a Gammaproteobacterium of the family Morganellaceae, and is a lethal pathogen of insects. It lives in the gut of an entomopathogenic nematode of the family Heterorhabditidae. When the nematode infects an insect, ''P. luminescens'' is released into the blood stream and rapidly kills the insect host (within 48 hours) by producing toxins, such as the high molecular weight insecticidal protein complex Tca. ''P. luminescens'' also produces a proteic toxin through the expression of a single gene called ''makes caterpillars floppy'' (mcf). It also secretes enzymes which break down the body of the infected insect and bioconvert it into nutrients which can be used by both nematode and bacteria. In this way, both organisms gain enough nutrients to replicate (or reproduce in the case of the nematode) several times. The bacteria enter the nematode progeny as they develop. 3,5-Dihydroxy-4-isopropyl-trans-stilb ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Biology
Molecular biology is the branch of biology that seeks to understand the molecular basis of biological activity in and between cells, including biomolecular synthesis, modification, mechanisms, and interactions. The study of chemical and physical structure of biological macromolecules is known as molecular biology. Molecular biology was first described as an approach focused on the underpinnings of biological phenomena - uncovering the structures of biological molecules as well as their interactions, and how these interactions explain observations of classical biology. In 1945 the term molecular biology was used by physicist William Astbury. In 1953 Francis Crick, James Watson, Rosalind Franklin, and colleagues, working at Medical Research Council unit, Cavendish laboratory, Cambridge (now the MRC Laboratory of Molecular Biology), made a double helix model of DNA which changed the entire research scenario. They proposed the DNA structure based on previous research done by Ro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Methanocaldococcus Jannaschii
''Methanocaldococcus jannaschii'' (formerly ''Methanococcus jannaschii'') is a thermophilic methanogenic archaean in the class Methanococci. It was the first archaeon to have its complete genome sequenced. The sequencing identified many genes unique to the archaea. Many of the synthesis pathways for methanogenic cofactors were worked out biochemically in this organism, as were several other archaeal-specific metabolic pathways. History ''Methanocaldococcus jannaschii'' was isolated from a submarine hydrothermal vent at Woods Hole Oceanographic Institution. Sequencing ''Methanocaldococcus jannaschii'' was sequenced by a group at TIGR led by Craig Venter using whole-genome shotgun sequencing. ''Methanocaldococcus jannaschii'' represented the first member of the Archaea to have its genome sequenced. According to Venter, the unique features of the genome provided strong evidence that there are three domains of life. Taxonomy ''Methanocaldoccus jannaschii'' is a member o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Staphylococcus Aureus
''Staphylococcus aureus'' is a Gram-positive spherically shaped bacterium, a member of the Bacillota, and is a usual member of the microbiota of the body, frequently found in the upper respiratory tract and on the skin. It is often positive for catalase and nitrate reduction and is a facultative anaerobe that can grow without the need for oxygen. Although ''S. aureus'' usually acts as a commensal of the human microbiota, it can also become an opportunistic pathogen, being a common cause of skin infections including abscesses, respiratory infections such as sinusitis, and food poisoning. Pathogenic strains often promote infections by producing virulence factors such as potent protein toxins, and the expression of a cell-surface protein that binds and inactivates antibodies. ''S. aureus'' is one of the leading pathogens for deaths associated with antimicrobial resistance and the emergence of antibiotic-resistant strains, such as methicillin-resistant ''S. aureus'' (MRSA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Motif
In biology, a sequence motif is a nucleotide or amino-acid sequence pattern that is widespread and usually assumed to be related to biological function of the macromolecule. For example, an ''N''-glycosylation site motif can be defined as ''Asn, followed by anything but Pro, followed by either Ser or Thr, followed by anything but Pro residue''. Overview When a sequence motif appears in the exon of a gene, it may encode the "structural motif" of a protein; that is a stereotypical element of the overall structure of the protein. Nevertheless, motifs need not be associated with a distinctive secondary structure. " Noncoding" sequences are not translated into proteins, and nucleic acids with such motifs need not deviate from the typical shape (e.g. the "B-form" DNA double helix). Outside of gene exons, there exist regulatory sequence motifs and motifs within the " junk", such as satellite DNA. Some of these are believed to affect the shape of nucleic acids (see for example RN ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Translation (biology)
In molecular biology and genetics, translation is the process in which ribosomes in the cytoplasm or endoplasmic reticulum synthesize proteins after the process of transcription (biology), transcription of DNA to RNA in the cell's nucleus (cell), nucleus. The entire process is called gene expression. In translation, mRNA, messenger RNA (mRNA) is decoded in a ribosome, outside the nucleus, to produce a specific amino acid chain, or polypeptide. The polypeptide later protein folding, folds into an Activation energy, active protein and performs its functions in the Cell (biology), cell. The ribosome facilitates decoding by inducing the binding of Base pair, complementary tRNA anticodon sequences to mRNA codons. The tRNAs carry specific amino acids that are chained together into a polypeptide as the mRNA passes through and is "read" by the ribosome. Translation proceeds in three phases: # Initiation: The ribosome assembles around the target mRNA. The first tRNA is attached a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Antisense
In molecular biology and genetics, the sense of a nucleic acid molecule, particularly of a strand of DNA or RNA, refers to the nature of the roles of the strand and its complement in specifying a sequence of amino acids. Depending on the context, sense may have slightly different meanings. For example, negative-sense strand of DNA is equivalent to the template strand, whereas the positive-sense strand is the non-template strand whose nucleotide sequence is equivalent to the sequence of the mRNA transcript. DNA sense Because of the complementary nature of base-pairing between nucleic acid polymers, a double-stranded DNA molecule will be composed of two strands with sequences that are reverse complements of each other. To help molecular biologists specifically identify each strand individually, the two strands are usually differentiated as the "sense" strand and the "antisense" strand. An individual strand of DNA is referred to as positive-sense (also positive (+) or simply sense) i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
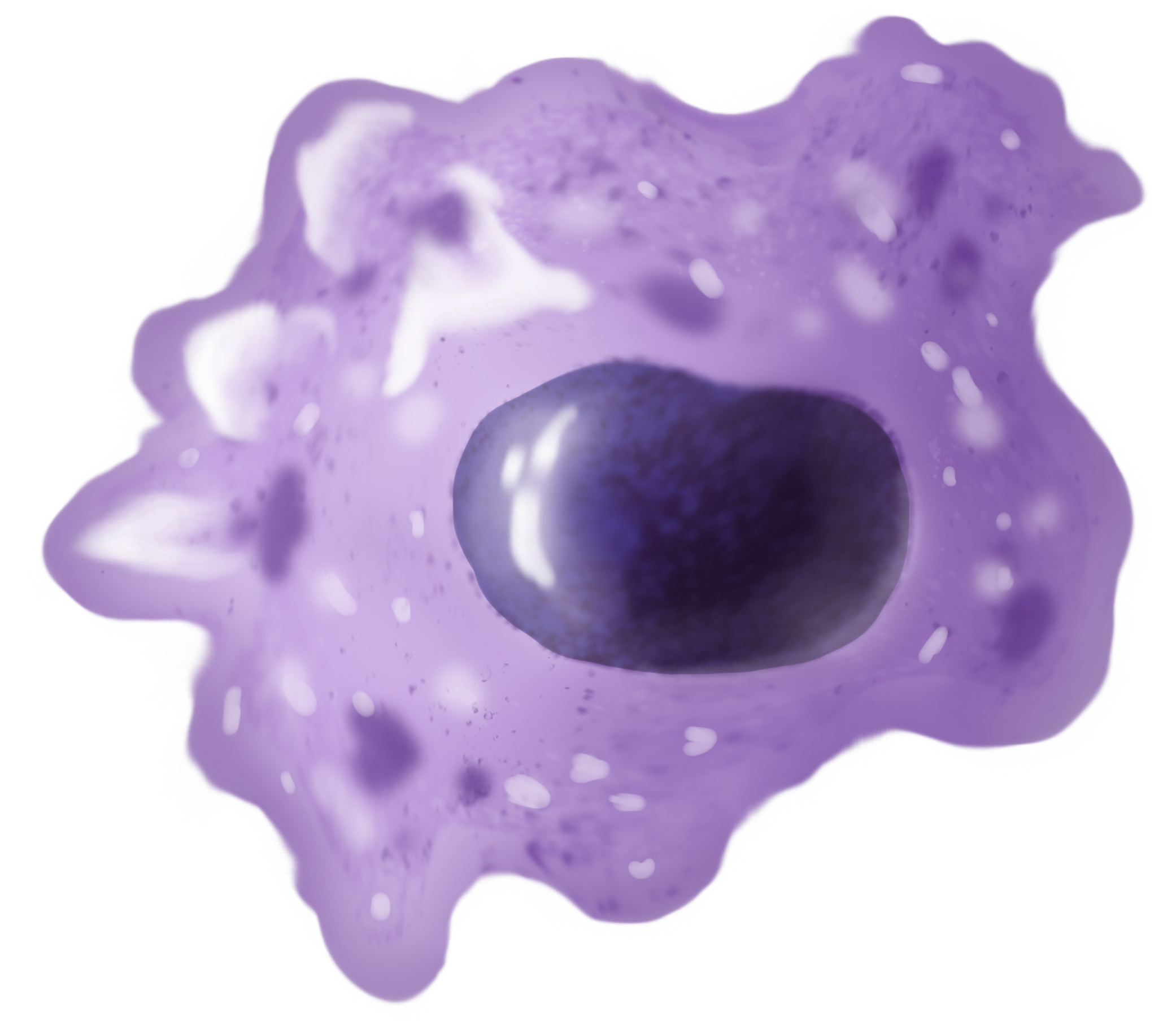
Macrophages
Macrophages (abbreviated as M φ, MΦ or MP) ( el, large eaters, from Greek ''μακρός'' (') = large, ''φαγεῖν'' (') = to eat) are a type of white blood cell of the immune system that engulfs and digests pathogens, such as cancer cells, microbes, cellular debris, and foreign substances, which do not have proteins that are specific to healthy body cells on their surface. The process is called phagocytosis, which acts to defend the host against infection and injury. These large phagocytes are found in essentially all tissues, where they patrol for potential pathogens by amoeboid movement. They take various forms (with various names) throughout the body (e.g., histiocytes, Kupffer cells, alveolar macrophages, microglia, and others), but all are part of the mononuclear phagocyte system. Besides phagocytosis, they play a critical role in nonspecific defense (innate immunity) and also help initiate specific defense mechanisms (adaptive immunity) by recruiting other immun ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |